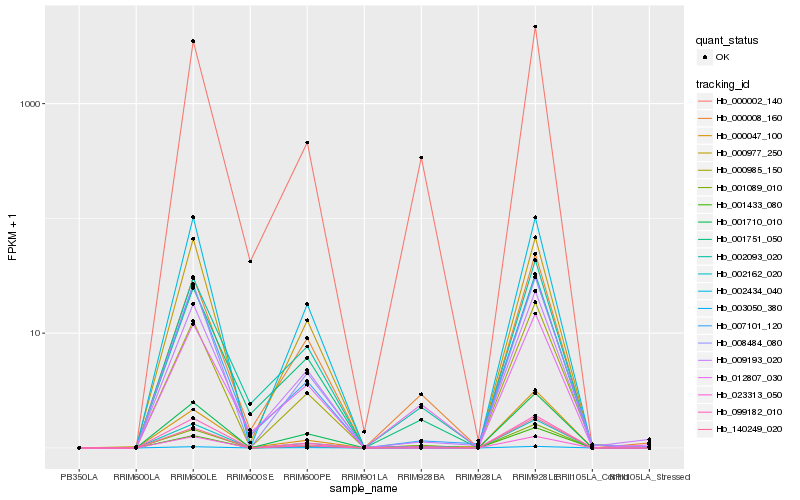
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002162_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_21655 [Jatropha curcas] |
| 2 |
Hb_000002_140 |
0.0678355482 |
- |
- |
hypothetical protein JCGZ_02318 [Jatropha curcas] |
| 3 |
Hb_001710_010 |
0.0693738463 |
- |
- |
Uncharacterized protein TCM_001832 [Theobroma cacao] |
| 4 |
Hb_140249_020 |
0.0749520262 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein CMB1-like [Jatropha curcas] |
| 5 |
Hb_000008_160 |
0.0874082179 |
- |
- |
carboxy-lyase, putative [Ricinus communis] |
| 6 |
Hb_001433_080 |
0.0991752156 |
- |
- |
Pectinesterase precursor, putative [Ricinus communis] |
| 7 |
Hb_001089_010 |
0.0992247409 |
- |
- |
PREDICTED: GDSL esterase/lipase 5-like [Jatropha curcas] |
| 8 |
Hb_001751_050 |
0.0998524316 |
- |
- |
PREDICTED: phytol kinase 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002093_020 |
0.105693159 |
- |
- |
hypothetical protein POPTR_0008s02960g [Populus trichocarpa] |
| 10 |
Hb_000047_100 |
0.1057164638 |
transcription factor |
TF Family: GRF |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_007101_120 |
0.1063207982 |
- |
- |
PREDICTED: uncharacterized protein LOC105649846 [Jatropha curcas] |
| 12 |
Hb_008484_080 |
0.1066504729 |
- |
- |
PREDICTED: crocetin glucosyltransferase, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_099182_010 |
0.1091532813 |
- |
- |
PREDICTED: uncharacterized protein LOC101214783 [Cucumis sativus] |
| 14 |
Hb_012807_030 |
0.112251279 |
- |
- |
PREDICTED: uncharacterized protein LOC105628042 isoform X1 [Jatropha curcas] |
| 15 |
Hb_023313_050 |
0.1134706221 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein rcd1-like [Jatropha curcas] |
| 16 |
Hb_003050_380 |
0.1146599157 |
- |
- |
hypothetical protein POPTR_0007s03380g [Populus trichocarpa] |
| 17 |
Hb_002434_040 |
0.1159541996 |
- |
- |
hypothetical protein CICLE_v10022672mg [Citrus clementina] |
| 18 |
Hb_009193_020 |
0.1161268595 |
- |
- |
PREDICTED: epoxide hydrolase 3 [Jatropha curcas] |
| 19 |
Hb_000985_150 |
0.1165552596 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 20 |
Hb_000977_250 |
0.1167308481 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |