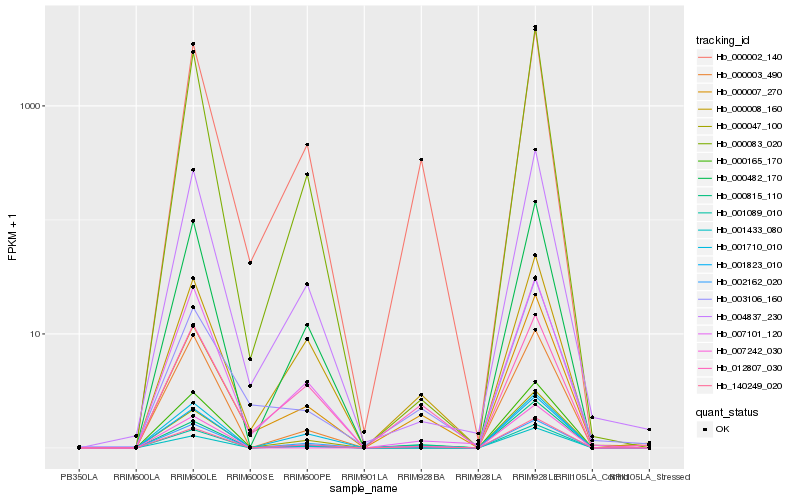
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001089_010 |
0.0 |
- |
- |
PREDICTED: GDSL esterase/lipase 5-like [Jatropha curcas] |
| 2 |
Hb_000002_140 |
0.0760744617 |
- |
- |
hypothetical protein JCGZ_02318 [Jatropha curcas] |
| 3 |
Hb_140249_020 |
0.093994309 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein CMB1-like [Jatropha curcas] |
| 4 |
Hb_002162_020 |
0.0992247409 |
- |
- |
hypothetical protein JCGZ_21655 [Jatropha curcas] |
| 5 |
Hb_000815_110 |
0.1069287874 |
- |
- |
RecName: Full=Casparian strip membrane protein 4; Short=RcCASP4 [Ricinus communis] |
| 6 |
Hb_001433_080 |
0.1169424227 |
- |
- |
Pectinesterase precursor, putative [Ricinus communis] |
| 7 |
Hb_000165_170 |
0.1178774524 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0010s19310g [Populus trichocarpa] |
| 8 |
Hb_000007_270 |
0.1198696103 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000047_100 |
0.1278322032 |
transcription factor |
TF Family: GRF |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007242_030 |
0.1432856014 |
- |
- |
PREDICTED: methylesterase 3-like isoform X2 [Tarenaya hassleriana] |
| 11 |
Hb_012807_030 |
0.1473506076 |
- |
- |
PREDICTED: uncharacterized protein LOC105628042 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000008_160 |
0.1476427928 |
- |
- |
carboxy-lyase, putative [Ricinus communis] |
| 13 |
Hb_003106_160 |
0.1536575712 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g67520 [Jatropha curcas] |
| 14 |
Hb_007101_120 |
0.1543818472 |
- |
- |
PREDICTED: uncharacterized protein LOC105649846 [Jatropha curcas] |
| 15 |
Hb_001823_010 |
0.1556304532 |
- |
- |
PREDICTED: ribosome-inactivating protein cucurmosin-like [Jatropha curcas] |
| 16 |
Hb_004837_230 |
0.1566423695 |
- |
- |
unknown [Populus trichocarpa] |
| 17 |
Hb_000083_020 |
0.1573319314 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 18 |
Hb_001710_010 |
0.1577699659 |
- |
- |
Uncharacterized protein TCM_001832 [Theobroma cacao] |
| 19 |
Hb_000003_490 |
0.1585732663 |
- |
- |
PREDICTED: uncharacterized protein LOC105631559 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000482_170 |
0.1591288692 |
transcription factor |
TF Family: C2C2-GATA |
GATA nirate-inducible carbon-metabolism involved protein [Populus nigra x Populus x canadensis] |