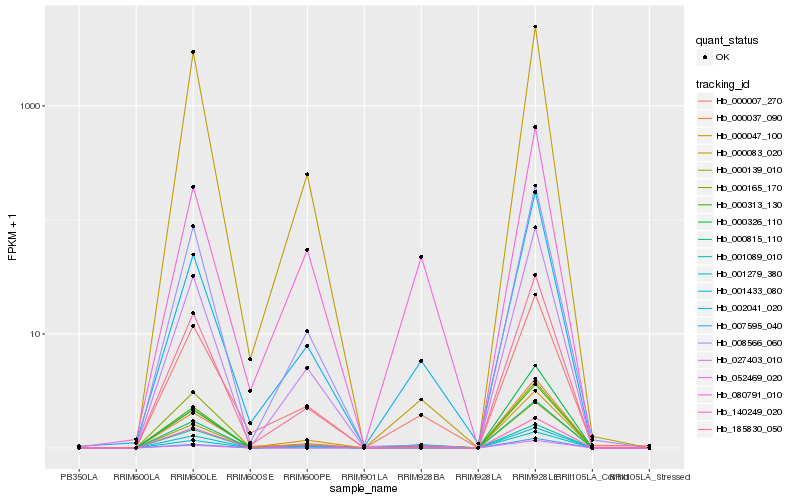
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000815_110 |
0.0 |
- |
- |
RecName: Full=Casparian strip membrane protein 4; Short=RcCASP4 [Ricinus communis] |
| 2 |
Hb_140249_020 |
0.0850488205 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein CMB1-like [Jatropha curcas] |
| 3 |
Hb_001433_080 |
0.085787911 |
- |
- |
Pectinesterase precursor, putative [Ricinus communis] |
| 4 |
Hb_000007_270 |
0.0955858899 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000047_100 |
0.0958191094 |
transcription factor |
TF Family: GRF |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002041_020 |
0.1011521633 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 7 |
Hb_001089_010 |
0.1069287874 |
- |
- |
PREDICTED: GDSL esterase/lipase 5-like [Jatropha curcas] |
| 8 |
Hb_000037_090 |
0.1152254618 |
- |
- |
Na(+)/H(+) antiporter, putative [Ricinus communis] |
| 9 |
Hb_007595_040 |
0.1195886779 |
- |
- |
ribosomal protein L2 (chloroplast) [Berberis bealei] |
| 10 |
Hb_052469_020 |
0.1202269005 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Vitis vinifera] |
| 11 |
Hb_008566_060 |
0.1215701823 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_185830_050 |
0.1219535092 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 [Jatropha curcas] |
| 13 |
Hb_000326_110 |
0.1227958937 |
- |
- |
PREDICTED: uncharacterized protein LOC105632350 [Jatropha curcas] |
| 14 |
Hb_080791_010 |
0.1245702335 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 15 |
Hb_000313_130 |
0.1284943275 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_027403_010 |
0.1286878308 |
- |
- |
P66 protein [Hevea brasiliensis] |
| 17 |
Hb_001279_380 |
0.1287131813 |
- |
- |
PREDICTED: uncharacterized protein LOC105115824 isoform X1 [Populus euphratica] |
| 18 |
Hb_000165_170 |
0.128934033 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0010s19310g [Populus trichocarpa] |
| 19 |
Hb_000139_010 |
0.1298755436 |
- |
- |
- |
| 20 |
Hb_000083_020 |
0.1324014195 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |