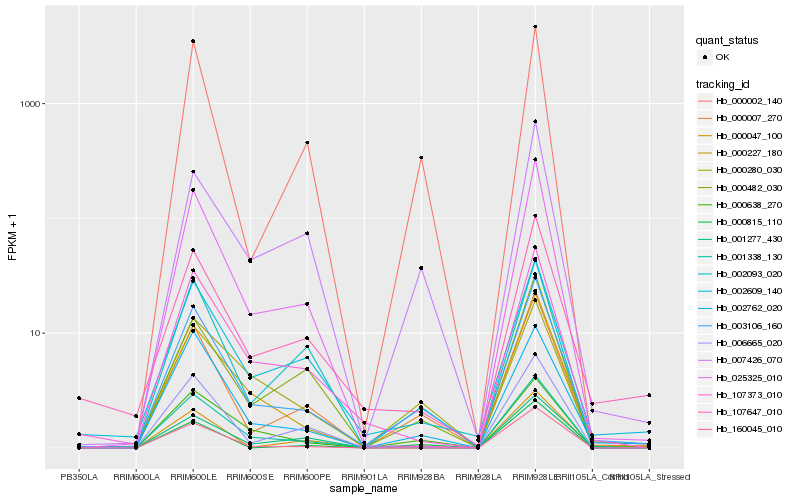
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003106_160 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g67520 [Jatropha curcas] |
| 2 |
Hb_000007_270 |
0.0794085043 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002762_020 |
0.112754209 |
- |
- |
hypothetical protein VITISV_043202 [Vitis vinifera] |
| 4 |
Hb_000638_270 |
0.1206251176 |
- |
- |
- |
| 5 |
Hb_025325_010 |
0.1223557589 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 6 |
Hb_002609_140 |
0.1260024382 |
- |
- |
PREDICTED: sodium-dependent phosphate transport protein 1, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_007426_070 |
0.1283840858 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 8 |
Hb_000047_100 |
0.1303290824 |
transcription factor |
TF Family: GRF |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_107373_010 |
0.1306694098 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_107647_010 |
0.13487762 |
- |
- |
PREDICTED: uncharacterized protein ycf39 [Jatropha curcas] |
| 11 |
Hb_000227_180 |
0.1358407551 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000482_030 |
0.1359108224 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER-like isoform X1 [Populus euphratica] |
| 13 |
Hb_000280_030 |
0.1361384921 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 55-like [Jatropha curcas] |
| 14 |
Hb_000002_140 |
0.1380277725 |
- |
- |
hypothetical protein JCGZ_02318 [Jatropha curcas] |
| 15 |
Hb_001338_130 |
0.1381283347 |
- |
- |
hypothetical protein MTR_4g129340 [Medicago truncatula] |
| 16 |
Hb_160045_010 |
0.1389936327 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000815_110 |
0.1396309892 |
- |
- |
RecName: Full=Casparian strip membrane protein 4; Short=RcCASP4 [Ricinus communis] |
| 18 |
Hb_001277_430 |
0.1427280114 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14 [Jatropha curcas] |
| 19 |
Hb_002093_020 |
0.1436106915 |
- |
- |
hypothetical protein POPTR_0008s02960g [Populus trichocarpa] |
| 20 |
Hb_006665_020 |
0.144524104 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |