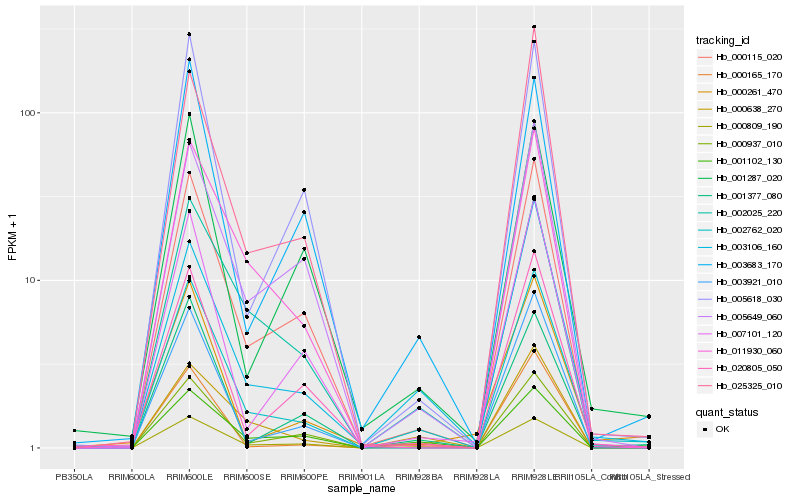
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002762_020 |
0.0 |
- |
- |
hypothetical protein VITISV_043202 [Vitis vinifera] |
| 2 |
Hb_000809_190 |
0.1051326663 |
- |
- |
PREDICTED: myosin-6-like [Jatropha curcas] |
| 3 |
Hb_003106_160 |
0.112754209 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g67520 [Jatropha curcas] |
| 4 |
Hb_000115_020 |
0.1129718289 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger family protein [Populus trichocarpa] |
| 5 |
Hb_002025_220 |
0.1219921243 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_000638_270 |
0.1272721217 |
- |
- |
- |
| 7 |
Hb_000165_170 |
0.1307899077 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0010s19310g [Populus trichocarpa] |
| 8 |
Hb_001102_130 |
0.1332762557 |
transcription factor |
TF Family: PHD |
PREDICTED: protein Jade-1 [Populus euphratica] |
| 9 |
Hb_003921_010 |
0.134764322 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_011930_060 |
0.1357031284 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 11 |
Hb_007101_120 |
0.135720239 |
- |
- |
PREDICTED: uncharacterized protein LOC105649846 [Jatropha curcas] |
| 12 |
Hb_020805_050 |
0.1362545638 |
- |
- |
PREDICTED: MLO-like protein 9 [Jatropha curcas] |
| 13 |
Hb_005649_060 |
0.1375577232 |
- |
- |
PREDICTED: uncharacterized protein LOC105644161 [Jatropha curcas] |
| 14 |
Hb_000937_010 |
0.1389725415 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005618_030 |
0.1425146922 |
- |
- |
PREDICTED: cytochrome P450 734A1-like [Jatropha curcas] |
| 16 |
Hb_001377_080 |
0.1437119667 |
- |
- |
PREDICTED: uncharacterized protein LOC105642864 [Jatropha curcas] |
| 17 |
Hb_000261_470 |
0.1449232839 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor FAMA isoform X2 [Populus euphratica] |
| 18 |
Hb_003683_170 |
0.1450749024 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001287_020 |
0.1453307141 |
- |
- |
SulA [Manihot esculenta] |
| 20 |
Hb_025325_010 |
0.1453776976 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |