| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
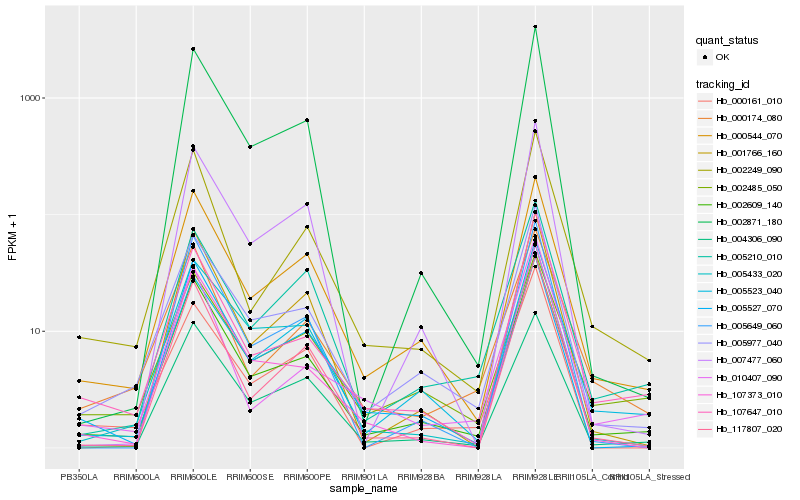
Hb_002609_140 |
0.0 |
- |
- |
PREDICTED: sodium-dependent phosphate transport protein 1, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_005977_040 |
0.0704654655 |
- |
- |
Photosystem I reaction center subunit IV A [Medicago truncatula] |
| 3 |
Hb_000544_070 |
0.0839519575 |
- |
- |
Peptidyl-prolyl cis-trans isomerase FKBP17-2 [Morus notabilis] |
| 4 |
Hb_002249_090 |
0.0876416054 |
- |
- |
PREDICTED: magnesium protoporphyrin IX methyltransferase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_004306_090 |
0.0876817986 |
- |
- |
PREDICTED: uncharacterized protein LOC105644392 isoform X2 [Jatropha curcas] |
| 6 |
Hb_107647_010 |
0.088264581 |
- |
- |
PREDICTED: uncharacterized protein ycf39 [Jatropha curcas] |
| 7 |
Hb_117807_020 |
0.0936080369 |
- |
- |
methyltransferase, putative [Ricinus communis] |
| 8 |
Hb_107373_010 |
0.0979395677 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_002871_180 |
0.0999166319 |
- |
- |
photosystem II 10 kDa polypeptide, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001766_160 |
0.1068268134 |
- |
- |
Serine/threonine-protein kinase SAPK1 [Gossypium arboreum] |
| 11 |
Hb_005649_060 |
0.1069747439 |
- |
- |
PREDICTED: uncharacterized protein LOC105644161 [Jatropha curcas] |
| 12 |
Hb_005523_040 |
0.1101445171 |
- |
- |
PREDICTED: uncharacterized protein LOC100253680 [Vitis vinifera] |
| 13 |
Hb_007477_060 |
0.1124823783 |
- |
- |
unknown [Medicago truncatula] |
| 14 |
Hb_005210_010 |
0.1135731633 |
- |
- |
PREDICTED: uncharacterized protein LOC105638163 [Jatropha curcas] |
| 15 |
Hb_005433_020 |
0.1163313283 |
- |
- |
PREDICTED: multicopper oxidase LPR1 [Jatropha curcas] |
| 16 |
Hb_010407_090 |
0.1167537066 |
- |
- |
PREDICTED: aldo-keto reductase isoform X1 [Jatropha curcas] |
| 17 |
Hb_000174_080 |
0.1189081426 |
- |
- |
4-nitrophenylphosphatase, putative [Ricinus communis] |
| 18 |
Hb_002485_050 |
0.1189535152 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 12, chloroplastic [Jatropha curcas] |
| 19 |
Hb_005527_070 |
0.1191799894 |
- |
- |
short-chain dehydrogenase/reductase family protein [Populus trichocarpa] |
| 20 |
Hb_000161_010 |
0.1209107133 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |