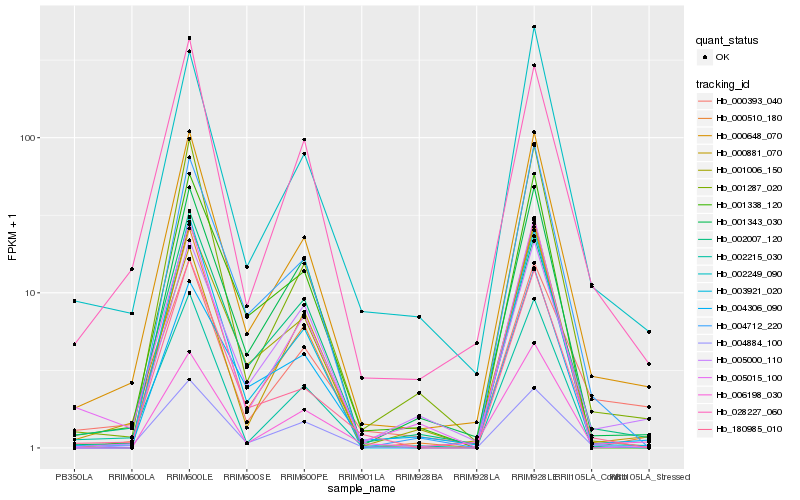
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000648_070 |
0.0 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 2 |
Hb_001287_020 |
0.08534614 |
- |
- |
SulA [Manihot esculenta] |
| 3 |
Hb_002249_090 |
0.0959121544 |
- |
- |
PREDICTED: magnesium protoporphyrin IX methyltransferase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000510_180 |
0.0963591076 |
- |
- |
unknown [Lotus japonicus] |
| 5 |
Hb_002215_030 |
0.1027697315 |
- |
- |
PREDICTED: uncharacterized protein LOC105640840 isoform X1 [Jatropha curcas] |
| 6 |
Hb_028227_060 |
0.1037190407 |
- |
- |
PREDICTED: non-specific lipid transfer protein GPI-anchored 1-like [Populus euphratica] |
| 7 |
Hb_002007_120 |
0.1050696374 |
- |
- |
PREDICTED: psbP domain-containing protein 7, chloroplastic [Jatropha curcas] |
| 8 |
Hb_004884_100 |
0.1058179287 |
- |
- |
PREDICTED: lectin-domain containing receptor kinase VI.4-like [Jatropha curcas] |
| 9 |
Hb_000881_070 |
0.1062957082 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_005015_100 |
0.1071367502 |
- |
- |
PREDICTED: uncharacterized protein LOC105636541 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001343_030 |
0.1075695472 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit N, chloroplastic [Jatropha curcas] |
| 12 |
Hb_006198_030 |
0.1113023208 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004306_090 |
0.1126056931 |
- |
- |
PREDICTED: uncharacterized protein LOC105644392 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001006_150 |
0.1140609854 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_000393_040 |
0.114250926 |
- |
- |
PREDICTED: uncharacterized protein LOC105128208 [Populus euphratica] |
| 16 |
Hb_001338_120 |
0.1144617129 |
- |
- |
hypothetical chloroplast RF68 (chloroplast) [Ipomoea batatas] |
| 17 |
Hb_005000_110 |
0.1146974407 |
- |
- |
PREDICTED: uncharacterized protein LOC105637872 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003921_020 |
0.1148382987 |
- |
- |
hypothetical protein JCGZ_10628 [Jatropha curcas] |
| 19 |
Hb_180985_010 |
0.1151372702 |
- |
- |
NDH-dependent cyclic electron flow 1 isoform 1 [Theobroma cacao] |
| 20 |
Hb_004712_220 |
0.115367665 |
- |
- |
conserved hypothetical protein [Ricinus communis] |