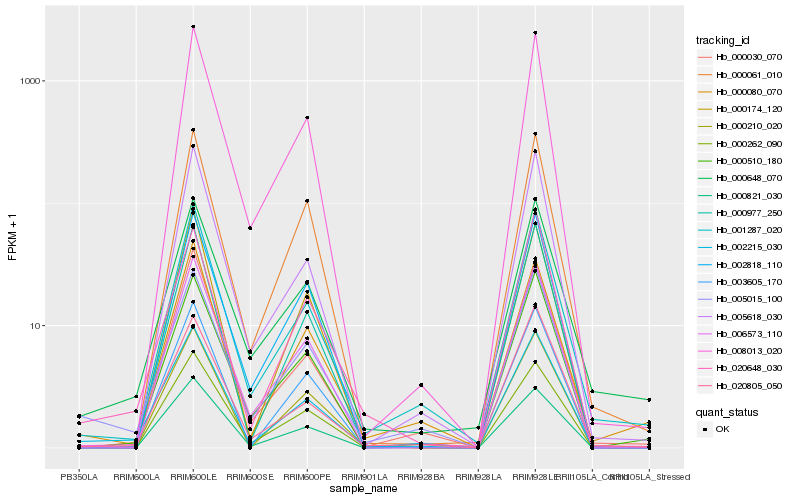
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002215_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640840 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005015_100 |
0.0743908162 |
- |
- |
PREDICTED: uncharacterized protein LOC105636541 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001287_020 |
0.0886970355 |
- |
- |
SulA [Manihot esculenta] |
| 4 |
Hb_020648_030 |
0.099938337 |
- |
- |
PREDICTED: long-chain-alcohol oxidase FAO1 [Jatropha curcas] |
| 5 |
Hb_008013_020 |
0.1003897168 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000977_250 |
0.1008301503 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 7 |
Hb_005618_030 |
0.1026021625 |
- |
- |
PREDICTED: cytochrome P450 734A1-like [Jatropha curcas] |
| 8 |
Hb_000262_090 |
0.1026185704 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 9 |
Hb_000648_070 |
0.1027697315 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 10 |
Hb_000821_030 |
0.1029279218 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-2 [Jatropha curcas] |
| 11 |
Hb_000080_070 |
0.1045180192 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 12 |
Hb_003605_170 |
0.1050193088 |
- |
- |
hypothetical protein POPTR_0013s12020g [Populus trichocarpa] |
| 13 |
Hb_006573_110 |
0.1055864119 |
- |
- |
PREDICTED: uncharacterized protein PAM68-like [Jatropha curcas] |
| 14 |
Hb_000030_070 |
0.1059069419 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_020805_050 |
0.1059208801 |
- |
- |
PREDICTED: MLO-like protein 9 [Jatropha curcas] |
| 16 |
Hb_000510_180 |
0.1059621329 |
- |
- |
unknown [Lotus japonicus] |
| 17 |
Hb_000174_120 |
0.1066670577 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 18 |
Hb_000061_010 |
0.1075977991 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002818_110 |
0.1083274501 |
transcription factor |
TF Family: bHLH |
hypothetical protein RCOM_1343120 [Ricinus communis] |
| 20 |
Hb_000210_020 |
0.1087780229 |
transcription factor |
TF Family: OFP |
Ovate family protein 13, putative [Theobroma cacao] |