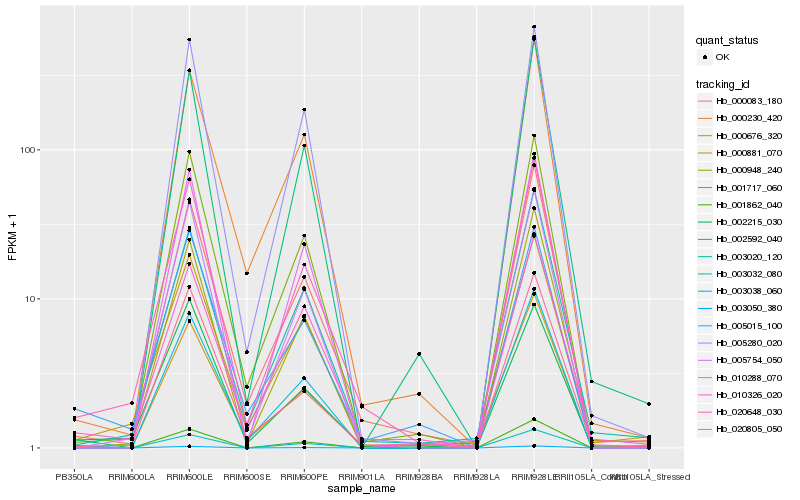
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020648_030 |
0.0 |
- |
- |
PREDICTED: long-chain-alcohol oxidase FAO1 [Jatropha curcas] |
| 2 |
Hb_000083_180 |
0.0732712159 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 3 |
Hb_001717_060 |
0.0775916679 |
- |
- |
PREDICTED: xylogen-like protein 11 [Gossypium raimondii] |
| 4 |
Hb_003038_060 |
0.0843969378 |
- |
- |
hypothetical protein POPTR_0007s13050g [Populus trichocarpa] |
| 5 |
Hb_005754_050 |
0.084714096 |
- |
- |
PREDICTED: uncharacterized protein LOC105636679 [Jatropha curcas] |
| 6 |
Hb_000881_070 |
0.0849291786 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_000948_240 |
0.0923678332 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003020_120 |
0.0925996402 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 9 |
Hb_010288_070 |
0.0970870517 |
transcription factor |
TF Family: MYB |
MYB-like DNA-binding domain protein [Theobroma cacao] |
| 10 |
Hb_003050_380 |
0.0980173356 |
- |
- |
hypothetical protein POPTR_0007s03380g [Populus trichocarpa] |
| 11 |
Hb_020805_050 |
0.0987841199 |
- |
- |
PREDICTED: MLO-like protein 9 [Jatropha curcas] |
| 12 |
Hb_000230_420 |
0.0993096726 |
- |
- |
GS [Hevea brasiliensis subsp. brasiliensis] |
| 13 |
Hb_002215_030 |
0.099938337 |
- |
- |
PREDICTED: uncharacterized protein LOC105640840 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003032_080 |
0.0999674946 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_010326_020 |
0.1004535943 |
- |
- |
PREDICTED: thioredoxin-like 4, chloroplastic [Jatropha curcas] |
| 16 |
Hb_005015_100 |
0.1010355425 |
- |
- |
PREDICTED: uncharacterized protein LOC105636541 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005280_020 |
0.1010449494 |
- |
- |
glycine cleavage system h protein, putative [Ricinus communis] |
| 18 |
Hb_001862_040 |
0.1010914407 |
- |
- |
heat shock protein 70 (HSP70)-interacting protein, putative [Ricinus communis] |
| 19 |
Hb_002592_040 |
0.1020205529 |
- |
- |
ACC oxidase 3 [Hevea brasiliensis] |
| 20 |
Hb_000676_320 |
0.1021979123 |
- |
- |
MADS-box transcription factor, partial [Clutia sp. DAV B80.252/F1980.8371] |