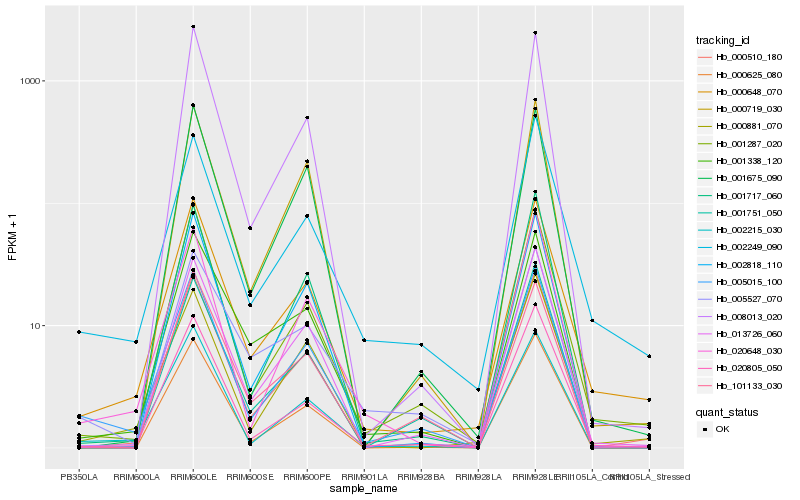
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005015_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636541 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002215_030 |
0.0743908162 |
- |
- |
PREDICTED: uncharacterized protein LOC105640840 isoform X1 [Jatropha curcas] |
| 3 |
Hb_020648_030 |
0.1010355425 |
- |
- |
PREDICTED: long-chain-alcohol oxidase FAO1 [Jatropha curcas] |
| 4 |
Hb_001287_020 |
0.1015236395 |
- |
- |
SulA [Manihot esculenta] |
| 5 |
Hb_000510_180 |
0.1017423846 |
- |
- |
unknown [Lotus japonicus] |
| 6 |
Hb_001717_060 |
0.1051110881 |
- |
- |
PREDICTED: xylogen-like protein 11 [Gossypium raimondii] |
| 7 |
Hb_000648_070 |
0.1071367502 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 8 |
Hb_013726_060 |
0.1092859845 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 9 |
Hb_002818_110 |
0.1108854216 |
transcription factor |
TF Family: bHLH |
hypothetical protein RCOM_1343120 [Ricinus communis] |
| 10 |
Hb_001338_120 |
0.1141163339 |
- |
- |
hypothetical chloroplast RF68 (chloroplast) [Ipomoea batatas] |
| 11 |
Hb_001751_050 |
0.1150824888 |
- |
- |
PREDICTED: phytol kinase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000625_080 |
0.1151058785 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 13 |
Hb_005527_070 |
0.1158787312 |
- |
- |
short-chain dehydrogenase/reductase family protein [Populus trichocarpa] |
| 14 |
Hb_002249_090 |
0.1163422678 |
- |
- |
PREDICTED: magnesium protoporphyrin IX methyltransferase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_008013_020 |
0.1165047269 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |
| 16 |
Hb_020805_050 |
0.1171959571 |
- |
- |
PREDICTED: MLO-like protein 9 [Jatropha curcas] |
| 17 |
Hb_101133_030 |
0.1180311441 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 18 |
Hb_000881_070 |
0.118211041 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_001675_090 |
0.11888816 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_000719_030 |
0.1192374289 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |