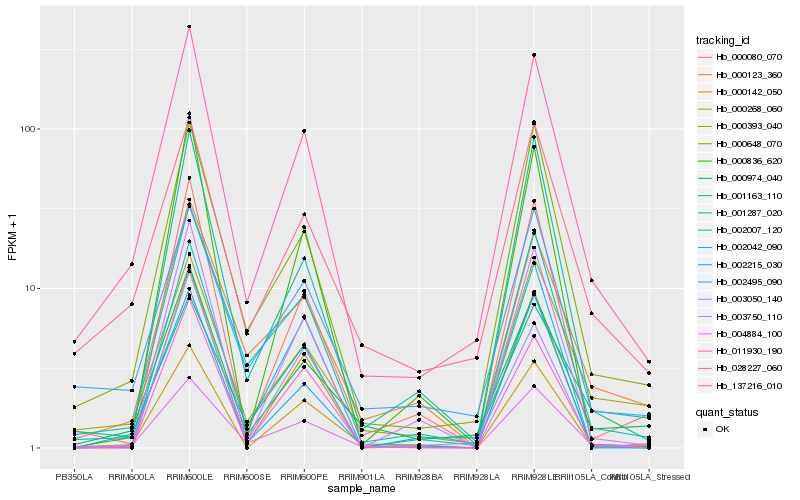
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028227_060 |
0.0 |
- |
- |
PREDICTED: non-specific lipid transfer protein GPI-anchored 1-like [Populus euphratica] |
| 2 |
Hb_000974_040 |
0.0942887884 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Jatropha curcas] |
| 3 |
Hb_002007_120 |
0.1025671444 |
- |
- |
PREDICTED: psbP domain-containing protein 7, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000648_070 |
0.1037190407 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 5 |
Hb_004884_100 |
0.1049793403 |
- |
- |
PREDICTED: lectin-domain containing receptor kinase VI.4-like [Jatropha curcas] |
| 6 |
Hb_000268_060 |
0.1128059692 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 7 |
Hb_137216_010 |
0.1151078911 |
- |
- |
PREDICTED: unknown protein DS12 from 2D-PAGE of leaf, chloroplastic-like isoform X2 [Citrus sinensis] |
| 8 |
Hb_002215_030 |
0.1233362611 |
- |
- |
PREDICTED: uncharacterized protein LOC105640840 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000080_070 |
0.1247996924 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000393_040 |
0.1311616913 |
- |
- |
PREDICTED: uncharacterized protein LOC105128208 [Populus euphratica] |
| 11 |
Hb_003750_110 |
0.1329899273 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |
| 12 |
Hb_002495_090 |
0.1333132374 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001287_020 |
0.1337030345 |
- |
- |
SulA [Manihot esculenta] |
| 14 |
Hb_011930_190 |
0.134868007 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 17 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000123_360 |
0.1394607613 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 1, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_002042_090 |
0.1399072353 |
- |
- |
PREDICTED: root phototropism protein 3-like [Jatropha curcas] |
| 17 |
Hb_003050_140 |
0.1419895482 |
- |
- |
PREDICTED: potassium channel KAT1 [Jatropha curcas] |
| 18 |
Hb_001163_110 |
0.1429290396 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PKL2 [Jatropha curcas] |
| 19 |
Hb_000836_620 |
0.1430905311 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000142_050 |
0.1439850107 |
- |
- |
PREDICTED: beta-glucosidase 18-like isoform X1 [Populus euphratica] |