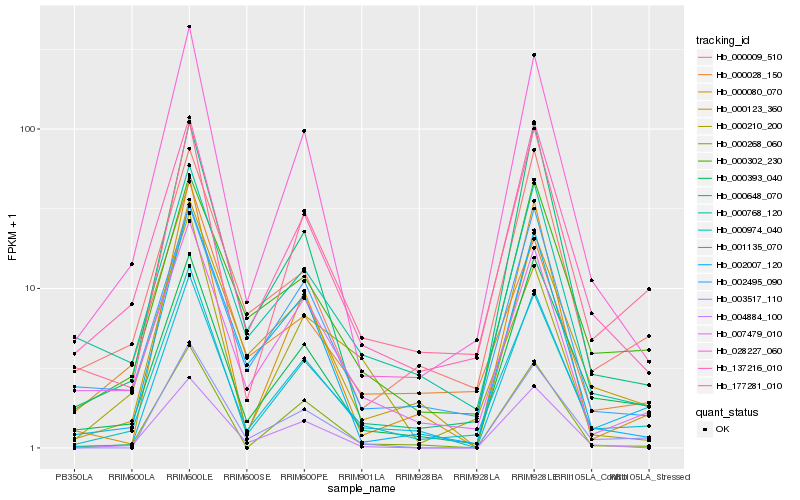
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000974_040 |
0.0 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Jatropha curcas] |
| 2 |
Hb_028227_060 |
0.0942887884 |
- |
- |
PREDICTED: non-specific lipid transfer protein GPI-anchored 1-like [Populus euphratica] |
| 3 |
Hb_177281_010 |
0.1069072382 |
- |
- |
PREDICTED: 50S ribosomal protein 5, chloroplastic-like [Citrus sinensis] |
| 4 |
Hb_137216_010 |
0.1095253352 |
- |
- |
PREDICTED: unknown protein DS12 from 2D-PAGE of leaf, chloroplastic-like isoform X2 [Citrus sinensis] |
| 5 |
Hb_001135_070 |
0.1095392505 |
- |
- |
glutathione-s-transferase omega, putative [Ricinus communis] |
| 6 |
Hb_000393_040 |
0.1130337588 |
- |
- |
PREDICTED: uncharacterized protein LOC105128208 [Populus euphratica] |
| 7 |
Hb_000123_360 |
0.1270751398 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 1, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_003517_110 |
0.1272878254 |
- |
- |
PREDICTED: uncharacterized protein LOC105633058 [Jatropha curcas] |
| 9 |
Hb_000268_060 |
0.1282569927 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 10 |
Hb_007479_010 |
0.1323934764 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 11 |
Hb_000648_070 |
0.1334403182 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 12 |
Hb_002495_090 |
0.1362141312 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000302_230 |
0.1369588644 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000080_070 |
0.1391984393 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000210_200 |
0.1420296147 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 16 |
Hb_002007_120 |
0.1420587876 |
- |
- |
PREDICTED: psbP domain-containing protein 7, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000028_150 |
0.1433690484 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 18 |
Hb_000768_120 |
0.1446031934 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigB [Jatropha curcas] |
| 19 |
Hb_004884_100 |
0.1470226187 |
- |
- |
PREDICTED: lectin-domain containing receptor kinase VI.4-like [Jatropha curcas] |
| 20 |
Hb_000009_510 |
0.1485166452 |
- |
- |
PREDICTED: uncharacterized protein LOC105126496 [Populus euphratica] |