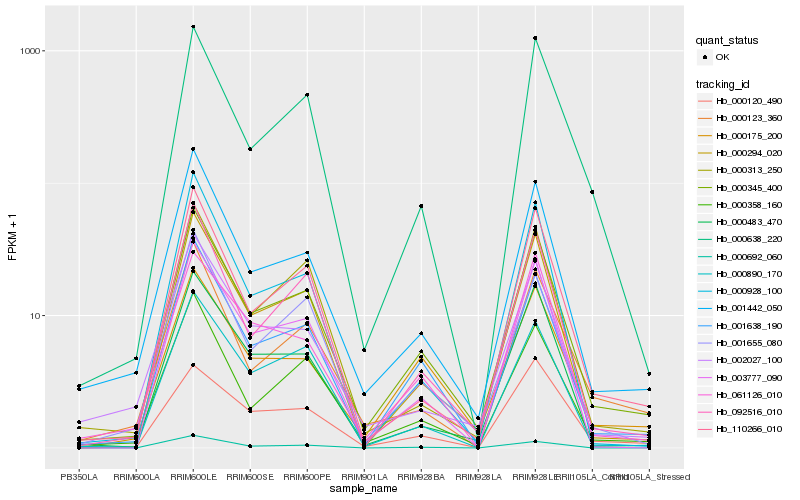
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000345_400 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001638_190 |
0.0714902139 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000294_020 |
0.0816514379 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 4 |
Hb_001442_050 |
0.0830329679 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit U, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000358_160 |
0.0914743598 |
- |
- |
PREDICTED: uncharacterized protein LOC105637769 isoform X1 [Jatropha curcas] |
| 6 |
Hb_092516_010 |
0.0923517659 |
- |
- |
hypothetical protein JCGZ_12248 [Jatropha curcas] |
| 7 |
Hb_110266_010 |
0.0948108619 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g21222-like [Jatropha curcas] |
| 8 |
Hb_000638_220 |
0.1036921487 |
- |
- |
PREDICTED: photosystem I reaction center subunit N, chloroplastic [Jatropha curcas] |
| 9 |
Hb_003777_090 |
0.105469906 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 10 |
Hb_000175_200 |
0.1055833353 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000483_470 |
0.1070559323 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_001655_080 |
0.1076361873 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 13 |
Hb_000123_360 |
0.1108089297 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 1, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_000313_250 |
0.1132233508 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 15 |
Hb_002027_100 |
0.1136275119 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000890_170 |
0.1140653112 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |
| 17 |
Hb_061126_010 |
0.117678423 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 18 |
Hb_000928_100 |
0.1185027378 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 19 |
Hb_000692_060 |
0.1199682878 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 20 |
Hb_000120_490 |
0.1208597897 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3-like [Jatropha curcas] |