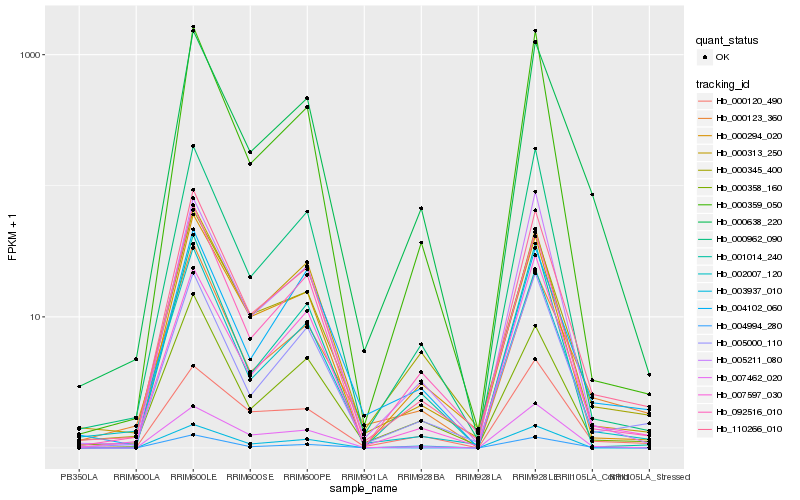
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000638_220 |
0.0 |
- |
- |
PREDICTED: photosystem I reaction center subunit N, chloroplastic [Jatropha curcas] |
| 2 |
Hb_110266_010 |
0.0911313716 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g21222-like [Jatropha curcas] |
| 3 |
Hb_000345_400 |
0.1036921487 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000962_090 |
0.1062410235 |
- |
- |
omega-6 fatty acid desaturase [Hevea brasiliensis] |
| 5 |
Hb_092516_010 |
0.1072754651 |
- |
- |
hypothetical protein JCGZ_12248 [Jatropha curcas] |
| 6 |
Hb_007462_020 |
0.1087007273 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 7 |
Hb_000123_360 |
0.1114330822 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 1, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_005000_110 |
0.1133129679 |
- |
- |
PREDICTED: uncharacterized protein LOC105637872 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005211_080 |
0.1137340911 |
- |
- |
hypothetical protein POPTR_0010s13740g [Populus trichocarpa] |
| 10 |
Hb_000313_250 |
0.1150562316 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 11 |
Hb_000294_020 |
0.1167892295 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 12 |
Hb_000358_160 |
0.1183888949 |
- |
- |
PREDICTED: uncharacterized protein LOC105637769 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004994_280 |
0.1187251717 |
- |
- |
hypothetical protein JCGZ_14421 [Jatropha curcas] |
| 14 |
Hb_002007_120 |
0.1198576629 |
- |
- |
PREDICTED: psbP domain-containing protein 7, chloroplastic [Jatropha curcas] |
| 15 |
Hb_007597_030 |
0.1204976691 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_004102_060 |
0.1218800279 |
- |
- |
JHL25H03.11 [Jatropha curcas] |
| 17 |
Hb_001014_240 |
0.1222916765 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.1 [Jatropha curcas] |
| 18 |
Hb_000120_490 |
0.1223277388 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3-like [Jatropha curcas] |
| 19 |
Hb_000359_050 |
0.1225272057 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_003937_010 |
0.122892776 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |