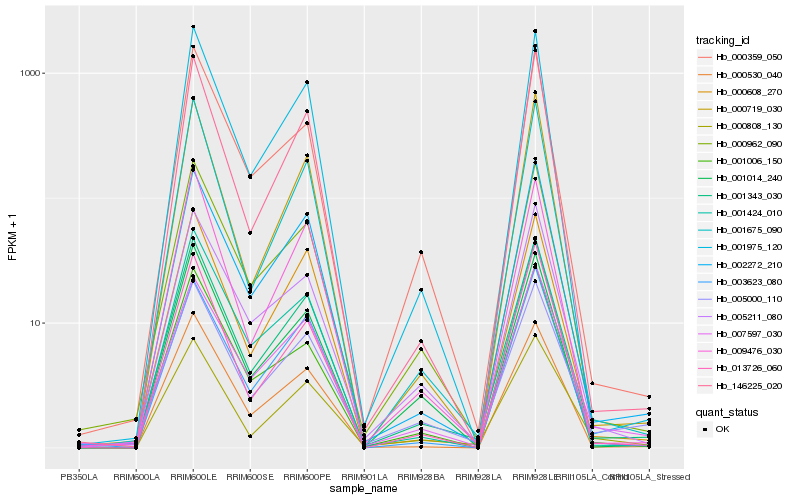
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001343_030 |
0.0 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit N, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000962_090 |
0.052305627 |
- |
- |
omega-6 fatty acid desaturase [Hevea brasiliensis] |
| 3 |
Hb_001975_120 |
0.0575845908 |
- |
- |
chlorophyll a/b binding protein type II [Glycine max] |
| 4 |
Hb_002272_210 |
0.0588057735 |
- |
- |
PREDICTED: UPF0603 protein At1g54780, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000530_040 |
0.0672261541 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005211_080 |
0.0677340234 |
- |
- |
hypothetical protein POPTR_0010s13740g [Populus trichocarpa] |
| 7 |
Hb_000359_050 |
0.0681296128 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_001675_090 |
0.0681440457 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_005000_110 |
0.0697772039 |
- |
- |
PREDICTED: uncharacterized protein LOC105637872 isoform X1 [Jatropha curcas] |
| 10 |
Hb_146225_020 |
0.073113145 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 11 |
Hb_000719_030 |
0.0736174781 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_007597_030 |
0.0743030357 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_000608_270 |
0.0758914771 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_009476_030 |
0.0759028113 |
- |
- |
fatty acid desaturase [Manihot esculenta] |
| 15 |
Hb_001014_240 |
0.0762619476 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.1 [Jatropha curcas] |
| 16 |
Hb_001424_010 |
0.0763460607 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 17 |
Hb_001006_150 |
0.0767791472 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_003623_080 |
0.0770404545 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Populus euphratica] |
| 19 |
Hb_013726_060 |
0.0787747172 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 20 |
Hb_000808_130 |
0.0798425285 |
- |
- |
3-5 exonuclease, putative [Ricinus communis] |