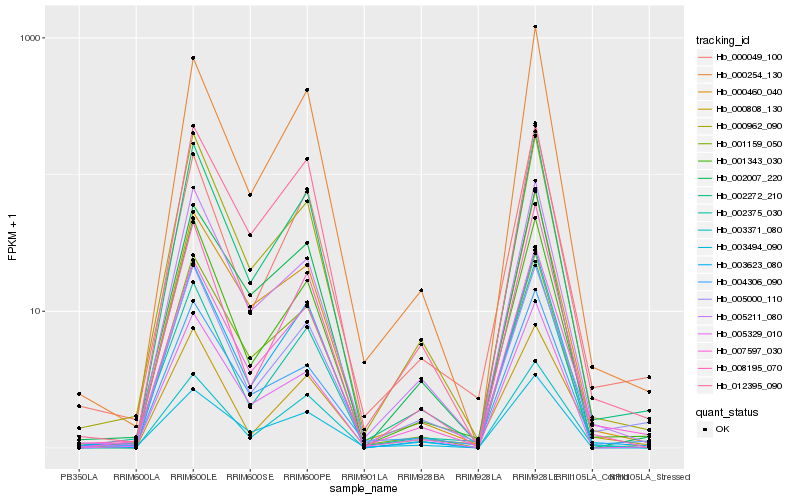
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007597_030 |
0.0 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_002272_210 |
0.0612843409 |
- |
- |
PREDICTED: UPF0603 protein At1g54780, chloroplastic [Jatropha curcas] |
| 3 |
Hb_005000_110 |
0.0718106496 |
- |
- |
PREDICTED: uncharacterized protein LOC105637872 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001343_030 |
0.0743030357 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit N, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000460_040 |
0.0763808311 |
- |
- |
PREDICTED: protein phosphatase 2C 57 [Jatropha curcas] |
| 6 |
Hb_003623_080 |
0.0780548017 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Populus euphratica] |
| 7 |
Hb_005211_080 |
0.0782322515 |
- |
- |
hypothetical protein POPTR_0010s13740g [Populus trichocarpa] |
| 8 |
Hb_000962_090 |
0.0816870591 |
- |
- |
omega-6 fatty acid desaturase [Hevea brasiliensis] |
| 9 |
Hb_000049_100 |
0.084802739 |
- |
- |
Photosystem II reaction center W protein, putative [Ricinus communis] |
| 10 |
Hb_004306_090 |
0.0865999719 |
- |
- |
PREDICTED: uncharacterized protein LOC105644392 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000254_130 |
0.0871176616 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_003371_080 |
0.087208578 |
- |
- |
hypothetical protein POPTR_0001s23660g [Populus trichocarpa] |
| 13 |
Hb_012395_090 |
0.0889897835 |
- |
- |
PREDICTED: protein PROTON GRADIENT REGULATION 5, chloroplastic [Jatropha curcas] |
| 14 |
Hb_005329_010 |
0.0894016699 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |
| 15 |
Hb_002375_030 |
0.0894194078 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 16 |
Hb_002007_220 |
0.0914591565 |
- |
- |
PREDICTED: signal recognition particle 43 kDa protein, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001159_050 |
0.0920108399 |
- |
- |
PREDICTED: uncharacterized protein LOC105645952 [Jatropha curcas] |
| 18 |
Hb_000808_130 |
0.0926659692 |
- |
- |
3-5 exonuclease, putative [Ricinus communis] |
| 19 |
Hb_003494_090 |
0.0938342507 |
transcription factor |
TF Family: MYB |
myb family transcription factor family protein [Populus trichocarpa] |
| 20 |
Hb_008195_070 |
0.0942951167 |
- |
- |
PREDICTED: thylakoid lumenal protein At1g12250, chloroplastic isoform X2 [Jatropha curcas] |