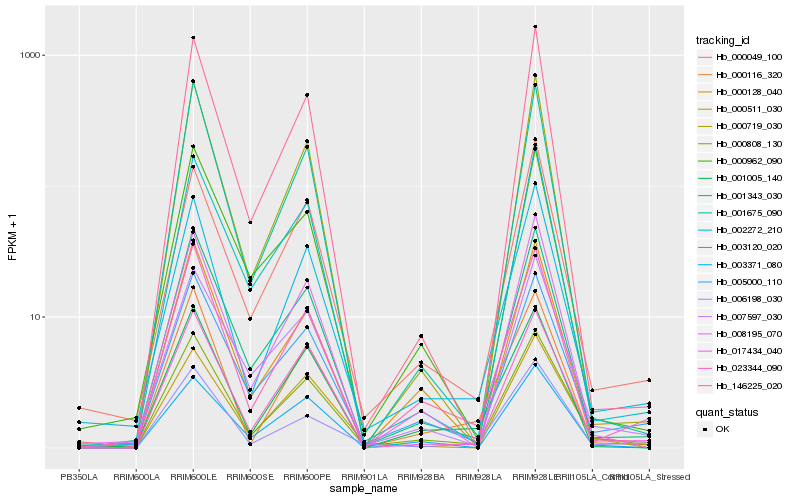
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000808_130 |
0.0 |
- |
- |
3-5 exonuclease, putative [Ricinus communis] |
| 2 |
Hb_005000_110 |
0.0728672027 |
- |
- |
PREDICTED: uncharacterized protein LOC105637872 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003120_020 |
0.0766164147 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 13-like [Prunus mume] |
| 4 |
Hb_001343_030 |
0.0798425285 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit N, chloroplastic [Jatropha curcas] |
| 5 |
Hb_017434_040 |
0.0851778031 |
- |
- |
PREDICTED: uncharacterized oxidoreductase At4g09670-like [Jatropha curcas] |
| 6 |
Hb_023344_090 |
0.0863527149 |
- |
- |
Photosystem II reaction center W protein, putative [Ricinus communis] |
| 7 |
Hb_007597_030 |
0.0926659692 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000049_100 |
0.0994055858 |
- |
- |
Photosystem II reaction center W protein, putative [Ricinus communis] |
| 9 |
Hb_000128_040 |
0.0996706914 |
- |
- |
PREDICTED: uncharacterized protein LOC105641228 isoform X1 [Jatropha curcas] |
| 10 |
Hb_008195_070 |
0.1004754229 |
- |
- |
PREDICTED: thylakoid lumenal protein At1g12250, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_000511_030 |
0.101286732 |
- |
- |
PREDICTED: uncharacterized protein LOC105636463 [Jatropha curcas] |
| 12 |
Hb_000116_320 |
0.1050237898 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 13 |
Hb_003371_080 |
0.1065445362 |
- |
- |
hypothetical protein POPTR_0001s23660g [Populus trichocarpa] |
| 14 |
Hb_002272_210 |
0.1066963609 |
- |
- |
PREDICTED: UPF0603 protein At1g54780, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001005_140 |
0.1073007454 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001675_090 |
0.1073143273 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000719_030 |
0.1073655576 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_000962_090 |
0.1082355325 |
- |
- |
omega-6 fatty acid desaturase [Hevea brasiliensis] |
| 19 |
Hb_006198_030 |
0.1091550726 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_146225_020 |
0.10915723 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |