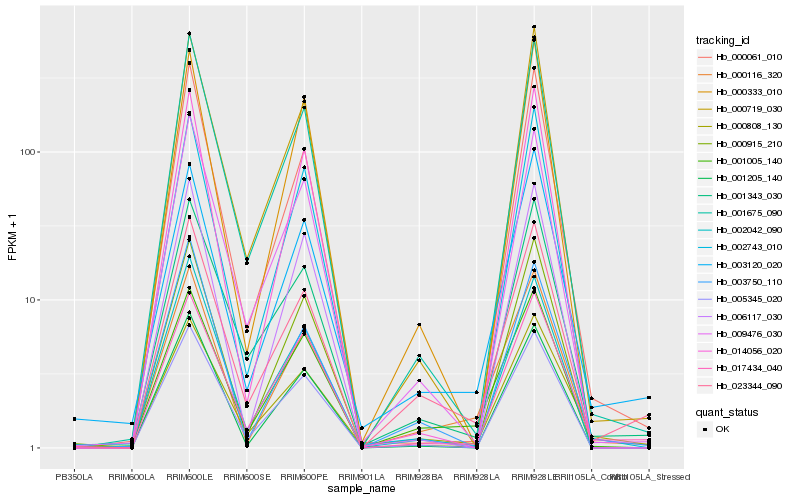
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000116_320 |
0.0 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 2 |
Hb_001005_140 |
0.0755736486 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_023344_090 |
0.0900263007 |
- |
- |
Photosystem II reaction center W protein, putative [Ricinus communis] |
| 4 |
Hb_003120_020 |
0.0925965999 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 13-like [Prunus mume] |
| 5 |
Hb_000808_130 |
0.1050237898 |
- |
- |
3-5 exonuclease, putative [Ricinus communis] |
| 6 |
Hb_000915_210 |
0.1139209871 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001675_090 |
0.1148239378 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_002042_090 |
0.1153012078 |
- |
- |
PREDICTED: root phototropism protein 3-like [Jatropha curcas] |
| 9 |
Hb_001205_140 |
0.1155030858 |
- |
- |
PREDICTED: cytochrome P450 90B1 [Jatropha curcas] |
| 10 |
Hb_000061_010 |
0.1179125363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_017434_040 |
0.1190012424 |
- |
- |
PREDICTED: uncharacterized oxidoreductase At4g09670-like [Jatropha curcas] |
| 12 |
Hb_003750_110 |
0.1201320722 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |
| 13 |
Hb_006117_030 |
0.1202727085 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_014056_020 |
0.1212139445 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_000333_010 |
0.1214368117 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |
| 16 |
Hb_009476_030 |
0.1217724452 |
- |
- |
fatty acid desaturase [Manihot esculenta] |
| 17 |
Hb_005345_020 |
0.1217736329 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 18 |
Hb_001343_030 |
0.1221565423 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit N, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002743_010 |
0.1222079287 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000719_030 |
0.1230610783 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |