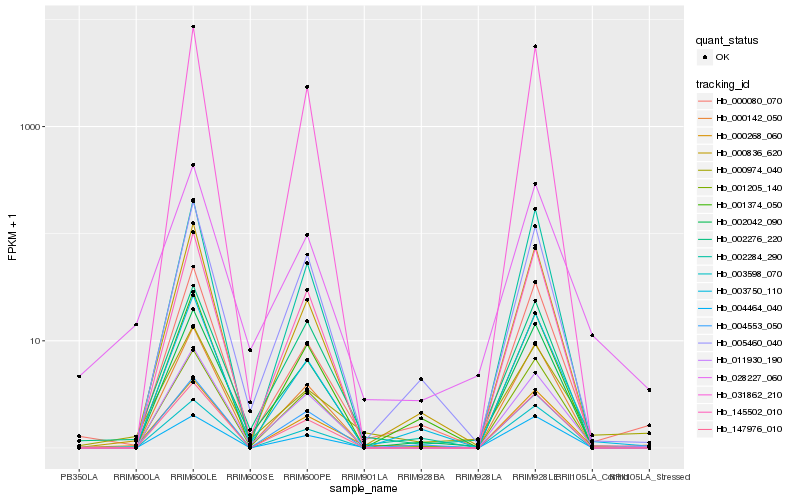
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000268_060 |
0.0 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 2 |
Hb_000080_070 |
0.0995880858 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 3 |
Hb_003750_110 |
0.1044431853 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |
| 4 |
Hb_028227_060 |
0.1128059692 |
- |
- |
PREDICTED: non-specific lipid transfer protein GPI-anchored 1-like [Populus euphratica] |
| 5 |
Hb_000836_620 |
0.1178836021 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_005460_040 |
0.1190105987 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 2 [Jatropha curcas] |
| 7 |
Hb_011930_190 |
0.1204054456 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 17 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000142_050 |
0.1215432364 |
- |
- |
PREDICTED: beta-glucosidase 18-like isoform X1 [Populus euphratica] |
| 9 |
Hb_001205_140 |
0.1227247557 |
- |
- |
PREDICTED: cytochrome P450 90B1 [Jatropha curcas] |
| 10 |
Hb_002284_290 |
0.1231424588 |
- |
- |
PREDICTED: transport and Golgi organization 2 homolog [Jatropha curcas] |
| 11 |
Hb_004553_050 |
0.1241767039 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g10020 [Jatropha curcas] |
| 12 |
Hb_004464_040 |
0.1241924506 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 13 |
Hb_001374_050 |
0.1246435232 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 14 |
Hb_147976_010 |
0.1257655294 |
- |
- |
- |
| 15 |
Hb_002042_090 |
0.1264317575 |
- |
- |
PREDICTED: root phototropism protein 3-like [Jatropha curcas] |
| 16 |
Hb_002276_220 |
0.1272429016 |
- |
- |
PREDICTED: uncharacterized protein LOC105640684 isoform X2 [Jatropha curcas] |
| 17 |
Hb_031862_210 |
0.1274650346 |
- |
- |
hypothetical protein JCGZ_24955 [Jatropha curcas] |
| 18 |
Hb_145502_010 |
0.1279809408 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB82-like [Jatropha curcas] |
| 19 |
Hb_003598_070 |
0.1279949811 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000974_040 |
0.1282569927 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Jatropha curcas] |