| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
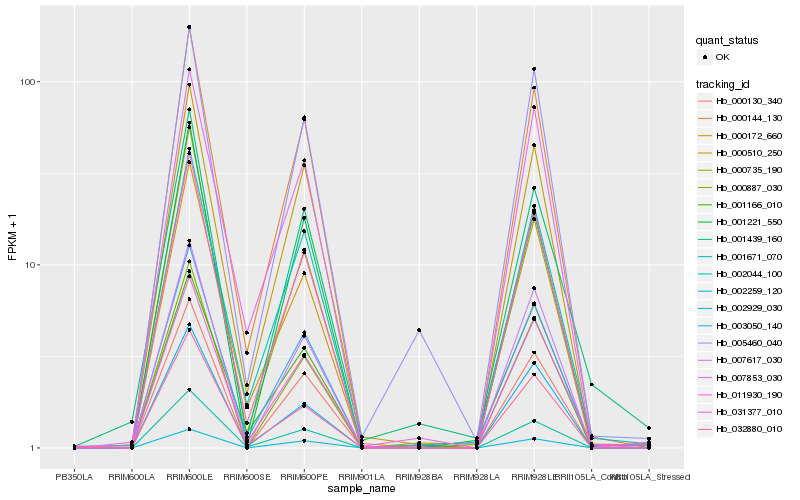
Hb_003050_140 |
0.0 |
- |
- |
PREDICTED: potassium channel KAT1 [Jatropha curcas] |
| 2 |
Hb_011930_190 |
0.0708502511 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 17 isoform X1 [Jatropha curcas] |
| 3 |
Hb_031377_010 |
0.0718279318 |
transcription factor |
TF Family: C2C2-YABBY |
unknown [Populus trichocarpa] |
| 4 |
Hb_000144_130 |
0.0758297022 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002044_100 |
0.0779844917 |
- |
- |
PREDICTED: uncharacterized protein LOC105643555 [Jatropha curcas] |
| 6 |
Hb_000510_250 |
0.0803394011 |
transcription factor |
TF Family: AUX/IAA |
auxin-induced protein aux28 [Populus trichocarpa] |
| 7 |
Hb_001671_070 |
0.0812107317 |
- |
- |
kinase, putative [Ricinus communis] |
| 8 |
Hb_001439_160 |
0.0816720815 |
- |
- |
PREDICTED: uncharacterized protein LOC105644097 [Jatropha curcas] |
| 9 |
Hb_001166_010 |
0.0842363072 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 10 |
Hb_000172_660 |
0.0902312317 |
- |
- |
flavonoid 3-hydroxylase, putative [Ricinus communis] |
| 11 |
Hb_032880_010 |
0.0902791663 |
- |
- |
PREDICTED: beta-glucosidase 24-like [Jatropha curcas] |
| 12 |
Hb_005460_040 |
0.090980942 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 2 [Jatropha curcas] |
| 13 |
Hb_000130_340 |
0.0922110296 |
- |
- |
- |
| 14 |
Hb_007617_030 |
0.0972924296 |
- |
- |
hypothetical protein JCGZ_26801 [Jatropha curcas] |
| 15 |
Hb_000735_190 |
0.0974293731 |
- |
- |
hypothetical protein POPTR_0006s19340g [Populus trichocarpa] |
| 16 |
Hb_001221_550 |
0.097622905 |
- |
- |
Corticosteroid 11-beta-dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_002929_030 |
0.0977466532 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 18 |
Hb_000887_030 |
0.0980558877 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 19 |
Hb_002259_120 |
0.0997674143 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 20 |
Hb_007853_030 |
0.101215189 |
- |
- |
structural molecule, putative [Ricinus communis] |