| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
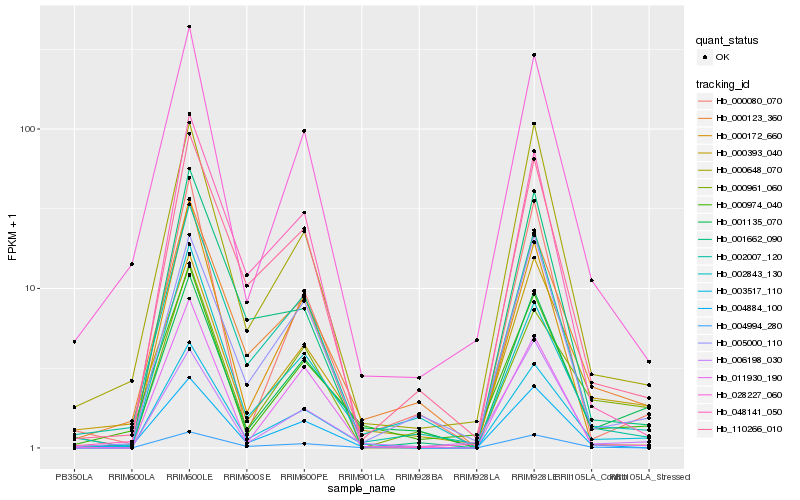
Hb_003517_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633058 [Jatropha curcas] |
| 2 |
Hb_001135_070 |
0.1022347404 |
- |
- |
glutathione-s-transferase omega, putative [Ricinus communis] |
| 3 |
Hb_004994_280 |
0.1219694512 |
- |
- |
hypothetical protein JCGZ_14421 [Jatropha curcas] |
| 4 |
Hb_000123_360 |
0.1237607801 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 1, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_000974_040 |
0.1272878254 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Jatropha curcas] |
| 6 |
Hb_110266_010 |
0.1345494781 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g21222-like [Jatropha curcas] |
| 7 |
Hb_004884_100 |
0.1362274103 |
- |
- |
PREDICTED: lectin-domain containing receptor kinase VI.4-like [Jatropha curcas] |
| 8 |
Hb_000393_040 |
0.138428063 |
- |
- |
PREDICTED: uncharacterized protein LOC105128208 [Populus euphratica] |
| 9 |
Hb_002007_120 |
0.1403901604 |
- |
- |
PREDICTED: psbP domain-containing protein 7, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000648_070 |
0.1414437926 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 11 |
Hb_011930_190 |
0.1468378566 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 17 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000080_070 |
0.1476138124 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000961_060 |
0.1479626473 |
- |
- |
uvb-resistance protein uvr8, putative [Ricinus communis] |
| 14 |
Hb_028227_060 |
0.1484287947 |
- |
- |
PREDICTED: non-specific lipid transfer protein GPI-anchored 1-like [Populus euphratica] |
| 15 |
Hb_002843_130 |
0.1488683443 |
- |
- |
PREDICTED: GRF1-interacting factor 2-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_005000_110 |
0.1498592918 |
- |
- |
PREDICTED: uncharacterized protein LOC105637872 isoform X1 [Jatropha curcas] |
| 17 |
Hb_048141_050 |
0.1502506049 |
- |
- |
hypothetical protein PRUPE_ppa009153mg [Prunus persica] |
| 18 |
Hb_001662_090 |
0.1522333542 |
- |
- |
PREDICTED: thiosulfate sulfurtransferase 16, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_006198_030 |
0.1541399581 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000172_660 |
0.1542954398 |
- |
- |
flavonoid 3-hydroxylase, putative [Ricinus communis] |