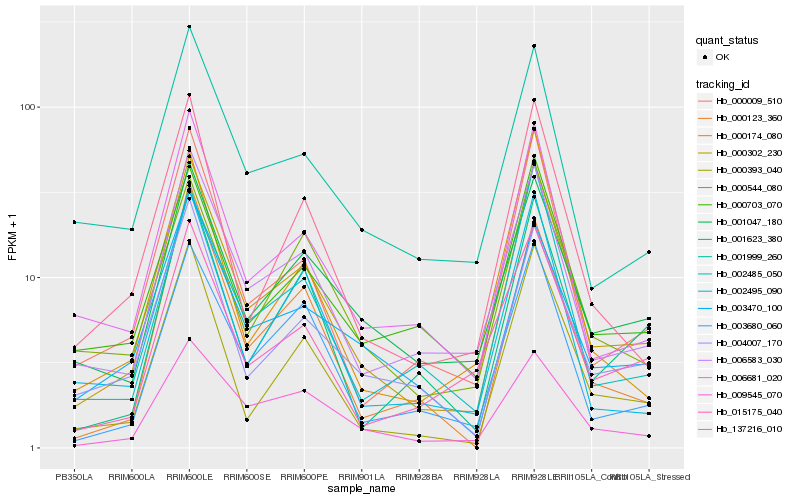
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000302_230 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000009_510 |
0.091575657 |
- |
- |
PREDICTED: uncharacterized protein LOC105126496 [Populus euphratica] |
| 3 |
Hb_000393_040 |
0.0959022861 |
- |
- |
PREDICTED: uncharacterized protein LOC105128208 [Populus euphratica] |
| 4 |
Hb_006583_030 |
0.1000799986 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like isoform X4 [Jatropha curcas] |
| 5 |
Hb_006681_020 |
0.1014215565 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP38, chloroplastic [Jatropha curcas] |
| 6 |
Hb_137216_010 |
0.1087563159 |
- |
- |
PREDICTED: unknown protein DS12 from 2D-PAGE of leaf, chloroplastic-like isoform X2 [Citrus sinensis] |
| 7 |
Hb_003680_060 |
0.1114033514 |
- |
- |
hypothetical protein VITISV_022322 [Vitis vinifera] |
| 8 |
Hb_002485_050 |
0.1157912766 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 12, chloroplastic [Jatropha curcas] |
| 9 |
Hb_004007_170 |
0.117048002 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 10 |
Hb_002495_090 |
0.1182845796 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 11 |
Hb_015175_040 |
0.121227905 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001047_180 |
0.1255098438 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_009545_070 |
0.1259502085 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g14820 [Jatropha curcas] |
| 14 |
Hb_000703_070 |
0.127644903 |
- |
- |
PREDICTED: uncharacterized protein LOC105635704 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003470_100 |
0.1278422024 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 16 |
Hb_000123_360 |
0.1278504103 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 1, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_000544_080 |
0.1281967611 |
- |
- |
PREDICTED: uncharacterized protein LOC105631062 isoform X3 [Jatropha curcas] |
| 18 |
Hb_001623_380 |
0.1282702258 |
- |
- |
PREDICTED: 7-hydroxymethyl chlorophyll a reductase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001999_260 |
0.1285222496 |
- |
- |
PREDICTED: 50S ribosomal protein L4, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000174_080 |
0.1290812529 |
- |
- |
4-nitrophenylphosphatase, putative [Ricinus communis] |