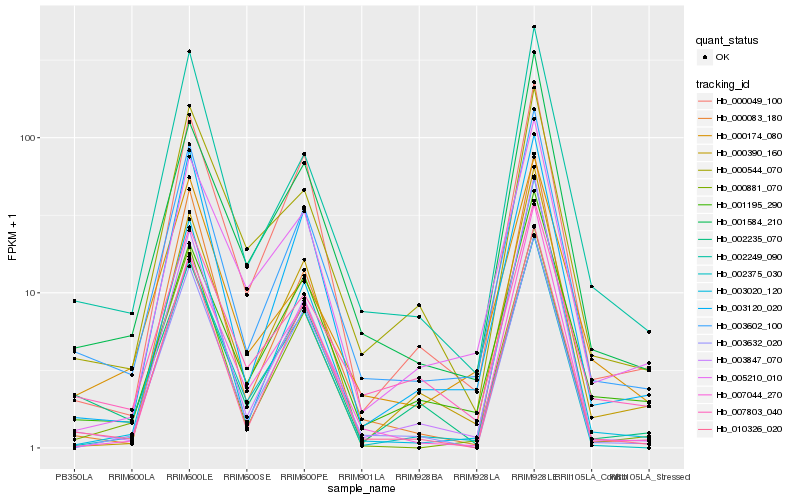
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003602_100 |
0.0 |
- |
- |
Glutamyl-tRNA reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_002249_090 |
0.0775554813 |
- |
- |
PREDICTED: magnesium protoporphyrin IX methyltransferase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001195_290 |
0.0932321689 |
- |
- |
PREDICTED: uncharacterized protein LOC105633792 [Jatropha curcas] |
| 4 |
Hb_003120_020 |
0.0934854264 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 13-like [Prunus mume] |
| 5 |
Hb_010326_020 |
0.0948137555 |
- |
- |
PREDICTED: thioredoxin-like 4, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000049_100 |
0.0977127456 |
- |
- |
Photosystem II reaction center W protein, putative [Ricinus communis] |
| 7 |
Hb_007803_040 |
0.1044718733 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP37, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000881_070 |
0.1047050131 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_000174_080 |
0.1053071275 |
- |
- |
4-nitrophenylphosphatase, putative [Ricinus communis] |
| 10 |
Hb_000390_160 |
0.1065827112 |
- |
- |
PREDICTED: adenosine kinase [Jatropha curcas] |
| 11 |
Hb_002235_070 |
0.1069374627 |
- |
- |
PREDICTED: uncharacterized protein LOC105644108 [Jatropha curcas] |
| 12 |
Hb_001584_210 |
0.1075832082 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa b, chloroplastic [Jatropha curcas] |
| 13 |
Hb_003020_120 |
0.1079665461 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 14 |
Hb_005210_010 |
0.109301787 |
- |
- |
PREDICTED: uncharacterized protein LOC105638163 [Jatropha curcas] |
| 15 |
Hb_003847_070 |
0.1111942933 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_003632_020 |
0.1112188465 |
- |
- |
calcium-binding family protein [Populus trichocarpa] |
| 17 |
Hb_000544_070 |
0.1139602878 |
- |
- |
Peptidyl-prolyl cis-trans isomerase FKBP17-2 [Morus notabilis] |
| 18 |
Hb_007044_270 |
0.1169642823 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002375_030 |
0.1170699262 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 20 |
Hb_000083_180 |
0.1172220914 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |