| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
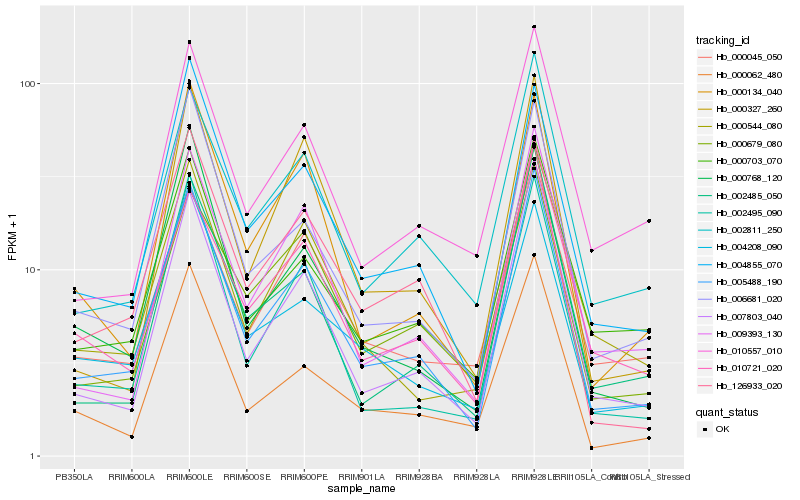
Hb_005488_190 |
0.0 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 2 |
Hb_007803_040 |
0.0795855222 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP37, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002811_250 |
0.0927398791 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_006681_020 |
0.0940168799 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP38, chloroplastic [Jatropha curcas] |
| 5 |
Hb_009393_130 |
0.0956685356 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X4 [Jatropha curcas] |
| 6 |
Hb_002495_090 |
0.0959240281 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000062_480 |
0.0974274877 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase-like 9 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004855_070 |
0.0976474885 |
- |
- |
PLASTID-SPECIFIC RIBOSOMAL protein 4 [Populus trichocarpa] |
| 9 |
Hb_002485_050 |
0.1005305546 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 12, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000703_070 |
0.1009166505 |
- |
- |
PREDICTED: uncharacterized protein LOC105635704 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000544_080 |
0.1072633735 |
- |
- |
PREDICTED: uncharacterized protein LOC105631062 isoform X3 [Jatropha curcas] |
| 12 |
Hb_000768_120 |
0.1078752529 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigB [Jatropha curcas] |
| 13 |
Hb_000679_080 |
0.109305562 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_010721_020 |
0.1097176538 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 4, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000134_040 |
0.1098304383 |
- |
- |
PREDICTED: 30S ribosomal protein 2, chloroplastic [Jatropha curcas] |
| 16 |
Hb_004208_090 |
0.1104712307 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 17 |
Hb_000045_050 |
0.1113674345 |
- |
- |
PREDICTED: probable zinc metalloprotease EGY2, chloroplastic [Jatropha curcas] |
| 18 |
Hb_126933_020 |
0.1118304222 |
- |
- |
PREDICTED: uncharacterized protein LOC105638525 [Jatropha curcas] |
| 19 |
Hb_010557_010 |
0.1133715342 |
- |
- |
PREDICTED: 50S ribosomal protein L3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000327_260 |
0.1144665111 |
- |
- |
conserved hypothetical protein [Ricinus communis] |