| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
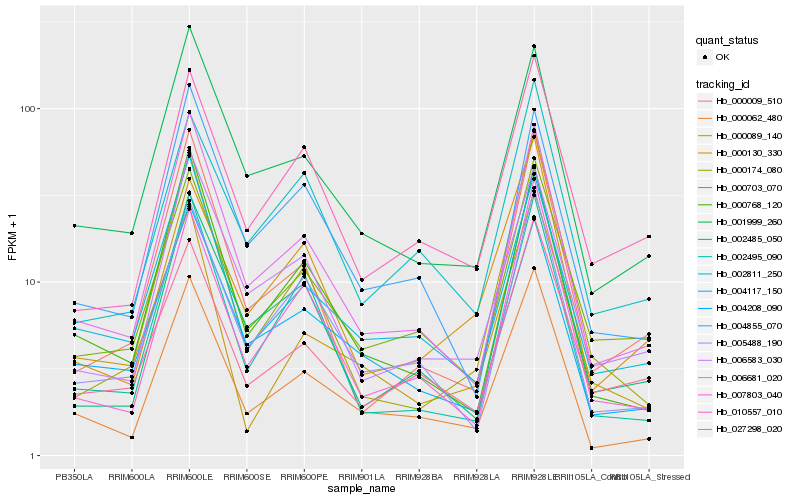
Hb_000062_480 |
0.0 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase-like 9 isoform X1 [Jatropha curcas] |
| 2 |
Hb_006681_020 |
0.0920058885 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP38, chloroplastic [Jatropha curcas] |
| 3 |
Hb_005488_190 |
0.0974274877 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 4 |
Hb_000768_120 |
0.1054624088 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigB [Jatropha curcas] |
| 5 |
Hb_007803_040 |
0.1094233984 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP37, chloroplastic [Jatropha curcas] |
| 6 |
Hb_004117_150 |
0.1125068002 |
- |
- |
PREDICTED: uncharacterized protein LOC105649105 [Jatropha curcas] |
| 7 |
Hb_004208_090 |
0.114641497 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 8 |
Hb_001999_260 |
0.1195298862 |
- |
- |
PREDICTED: 50S ribosomal protein L4, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002495_090 |
0.1214846156 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000703_070 |
0.1226027962 |
- |
- |
PREDICTED: uncharacterized protein LOC105635704 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002485_050 |
0.1271622723 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 12, chloroplastic [Jatropha curcas] |
| 12 |
Hb_006583_030 |
0.132280772 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like isoform X4 [Jatropha curcas] |
| 13 |
Hb_004855_070 |
0.1328679868 |
- |
- |
PLASTID-SPECIFIC RIBOSOMAL protein 4 [Populus trichocarpa] |
| 14 |
Hb_027298_020 |
0.1351400028 |
- |
- |
hypothetical protein POPTR_0005s05760g [Populus trichocarpa] |
| 15 |
Hb_000130_330 |
0.1360727141 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP3-like [Jatropha curcas] |
| 16 |
Hb_000009_510 |
0.1367735137 |
- |
- |
PREDICTED: uncharacterized protein LOC105126496 [Populus euphratica] |
| 17 |
Hb_002811_250 |
0.1372134116 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_010557_010 |
0.1373585903 |
- |
- |
PREDICTED: 50S ribosomal protein L3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000089_140 |
0.1414910784 |
- |
- |
PREDICTED: uncharacterized protein LOC105636682 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000174_080 |
0.1433896207 |
- |
- |
4-nitrophenylphosphatase, putative [Ricinus communis] |