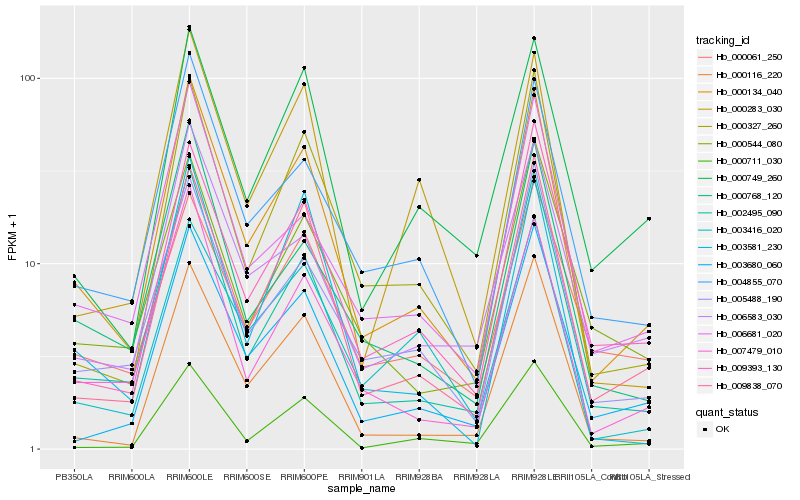
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000134_040 |
0.0 |
- |
- |
PREDICTED: 30S ribosomal protein 2, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002495_090 |
0.0890872631 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 3 |
Hb_004855_070 |
0.0967096481 |
- |
- |
PLASTID-SPECIFIC RIBOSOMAL protein 4 [Populus trichocarpa] |
| 4 |
Hb_009838_070 |
0.0972792441 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic [Jatropha curcas] |
| 5 |
Hb_007479_010 |
0.0985614877 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 6 |
Hb_003680_060 |
0.1063128375 |
- |
- |
hypothetical protein VITISV_022322 [Vitis vinifera] |
| 7 |
Hb_000327_260 |
0.1082336579 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005488_190 |
0.1098304383 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 9 |
Hb_000768_120 |
0.1122978007 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigB [Jatropha curcas] |
| 10 |
Hb_009393_130 |
0.1141256506 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X4 [Jatropha curcas] |
| 11 |
Hb_006583_030 |
0.1148558163 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like isoform X4 [Jatropha curcas] |
| 12 |
Hb_006681_020 |
0.1155532131 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP38, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000116_220 |
0.1162802972 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000749_260 |
0.1211506308 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000711_030 |
0.1290269022 |
- |
- |
hypothetical protein POPTR_0019s02860g [Populus trichocarpa] |
| 16 |
Hb_003581_230 |
0.1314514164 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SNF1-related protein kinase regulatory subunit gamma-1-like [Jatropha curcas] |
| 17 |
Hb_000061_250 |
0.1339486552 |
- |
- |
hypothetical protein JCGZ_11665 [Jatropha curcas] |
| 18 |
Hb_003416_020 |
0.1347602404 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 19 |
Hb_000544_080 |
0.1347630542 |
- |
- |
PREDICTED: uncharacterized protein LOC105631062 isoform X3 [Jatropha curcas] |
| 20 |
Hb_000283_030 |
0.1353838271 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA16-like [Populus euphratica] |