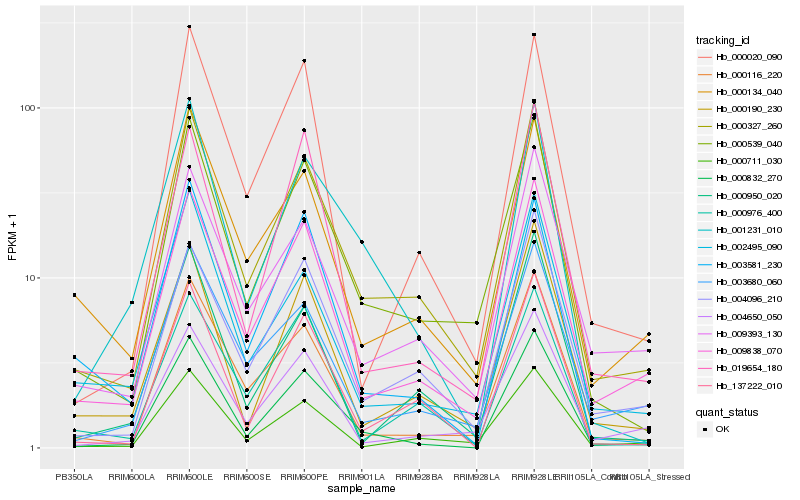
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000539_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636110 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000327_260 |
0.0708575214 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000116_220 |
0.1080927631 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_009838_070 |
0.1182537729 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000711_030 |
0.120367645 |
- |
- |
hypothetical protein POPTR_0019s02860g [Populus trichocarpa] |
| 6 |
Hb_000832_270 |
0.1218597464 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_137222_010 |
0.1287016561 |
- |
- |
PREDICTED: hippocampus abundant transcript-like protein 1 [Jatropha curcas] |
| 8 |
Hb_000190_230 |
0.1305537348 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 9 |
Hb_004096_210 |
0.1333337616 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |
| 10 |
Hb_004650_050 |
0.1338710764 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_001231_010 |
0.1376921423 |
- |
- |
calcium-binding family protein [Populus trichocarpa] |
| 12 |
Hb_002495_090 |
0.1378204302 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000134_040 |
0.1379074704 |
- |
- |
PREDICTED: 30S ribosomal protein 2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000020_090 |
0.1381785786 |
- |
- |
hypothetical protein JCGZ_09084 [Jatropha curcas] |
| 15 |
Hb_009393_130 |
0.1395801165 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X4 [Jatropha curcas] |
| 16 |
Hb_019654_180 |
0.1402390209 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 17 |
Hb_003680_060 |
0.1418415708 |
- |
- |
hypothetical protein VITISV_022322 [Vitis vinifera] |
| 18 |
Hb_003581_230 |
0.1424734639 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SNF1-related protein kinase regulatory subunit gamma-1-like [Jatropha curcas] |
| 19 |
Hb_000950_020 |
0.1443337332 |
- |
- |
PREDICTED: glycerol-3-phosphate dehydrogenase [NAD(+)] GPDHC1, cytosolic [Jatropha curcas] |
| 20 |
Hb_000976_400 |
0.145147125 |
- |
- |
synaptotagmin, putative [Ricinus communis] |