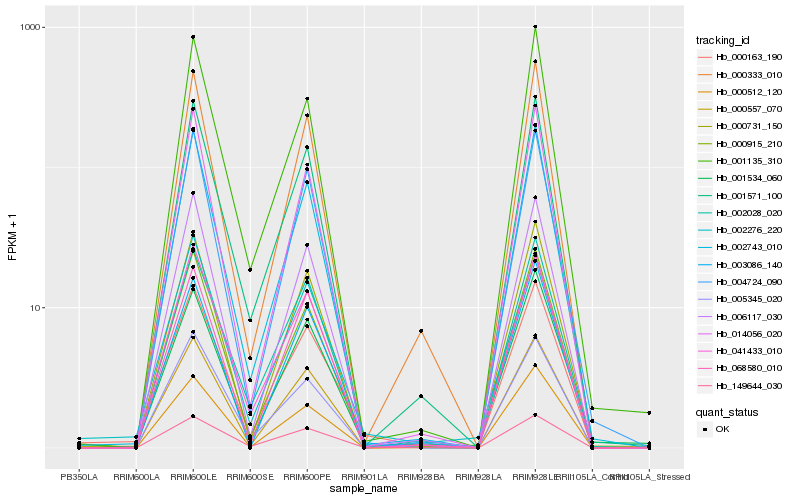
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000163_190 |
0.0 |
- |
- |
PREDICTED: 21 kDa protein [Populus euphratica] |
| 2 |
Hb_000915_210 |
0.0742270225 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_149644_030 |
0.0745796479 |
- |
- |
hypothetical protein POPTR_0005s18750g [Populus trichocarpa] |
| 4 |
Hb_002743_010 |
0.077542096 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001571_100 |
0.0805157256 |
- |
- |
chlorophyll A-B binding family protein [Populus trichocarpa] |
| 6 |
Hb_000731_150 |
0.0844437391 |
- |
- |
PREDICTED: cytochrome P450 77A1 [Jatropha curcas] |
| 7 |
Hb_000557_070 |
0.084962025 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 8 |
Hb_000333_010 |
0.0878067463 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |
| 9 |
Hb_006117_030 |
0.0887594273 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_014056_020 |
0.0895681615 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_002276_220 |
0.0905311682 |
- |
- |
PREDICTED: uncharacterized protein LOC105640684 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000512_120 |
0.092731701 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor NGA1-like [Jatropha curcas] |
| 13 |
Hb_041433_010 |
0.0936073415 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_002028_020 |
0.0938275217 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 15 |
Hb_004724_090 |
0.0939935603 |
- |
- |
PREDICTED: uncharacterized protein LOC105638638 [Jatropha curcas] |
| 16 |
Hb_005345_020 |
0.0958180089 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 17 |
Hb_003086_140 |
0.0968626945 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: protein YABBY 6-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_001135_310 |
0.0971223508 |
- |
- |
Chlorophyll a-b binding protein 6A [Morus notabilis] |
| 19 |
Hb_068580_010 |
0.0980336376 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 isoform X2 [Populus euphratica] |
| 20 |
Hb_001534_060 |
0.0984992532 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C3 [Jatropha curcas] |