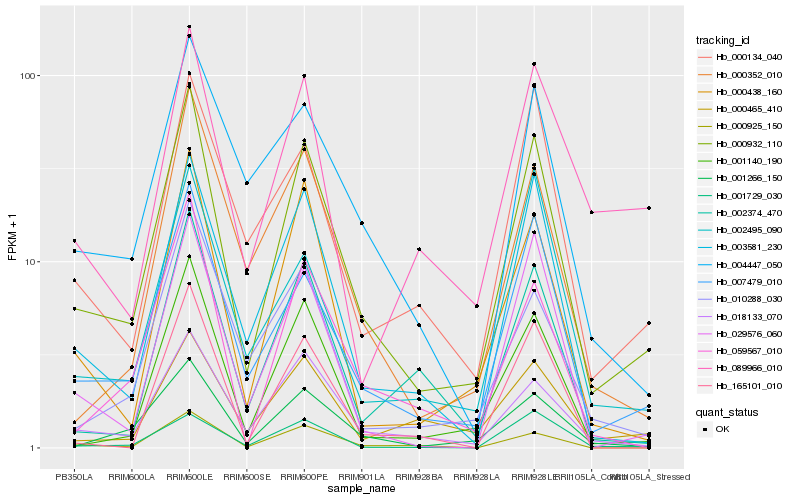
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000932_110 |
0.0 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 2 |
Hb_007479_010 |
0.1167517756 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 3 |
Hb_001140_190 |
0.1327462926 |
- |
- |
PREDICTED: A-kinase anchor protein 9 [Jatropha curcas] |
| 4 |
Hb_018133_070 |
0.1373699032 |
- |
- |
PREDICTED: RING-H2 finger protein ATL43 [Jatropha curcas] |
| 5 |
Hb_003581_230 |
0.1383669594 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SNF1-related protein kinase regulatory subunit gamma-1-like [Jatropha curcas] |
| 6 |
Hb_000438_160 |
0.1401725185 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_029576_060 |
0.1455455029 |
- |
- |
PREDICTED: uncharacterized protein LOC105646366 [Jatropha curcas] |
| 8 |
Hb_001266_150 |
0.146350749 |
- |
- |
hypothetical protein JCGZ_14961 [Jatropha curcas] |
| 9 |
Hb_000925_150 |
0.1514551132 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 10 |
Hb_000352_010 |
0.1577381158 |
- |
- |
PREDICTED: nudix hydrolase 13, mitochondrial [Jatropha curcas] |
| 11 |
Hb_059567_010 |
0.1596629879 |
- |
- |
PREDICTED: transcription factor PAR2-like [Jatropha curcas] |
| 12 |
Hb_002374_470 |
0.1610896448 |
- |
- |
PREDICTED: filament-like plant protein isoform X2 [Jatropha curcas] |
| 13 |
Hb_000134_040 |
0.1614896085 |
- |
- |
PREDICTED: 30S ribosomal protein 2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001729_030 |
0.1652283199 |
- |
- |
hypothetical protein VITISV_044457 [Vitis vinifera] |
| 15 |
Hb_004447_050 |
0.1690925752 |
- |
- |
PREDICTED: high-light-induced protein, chloroplastic [Jatropha curcas] |
| 16 |
Hb_010288_030 |
0.1696466928 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 17 |
Hb_165101_010 |
0.1698096791 |
- |
- |
PREDICTED: putative wall-associated receptor kinase-like 16 [Nelumbo nucifera] |
| 18 |
Hb_002495_090 |
0.1734634038 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000465_410 |
0.1737209003 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_089966_010 |
0.174196807 |
- |
- |
- |