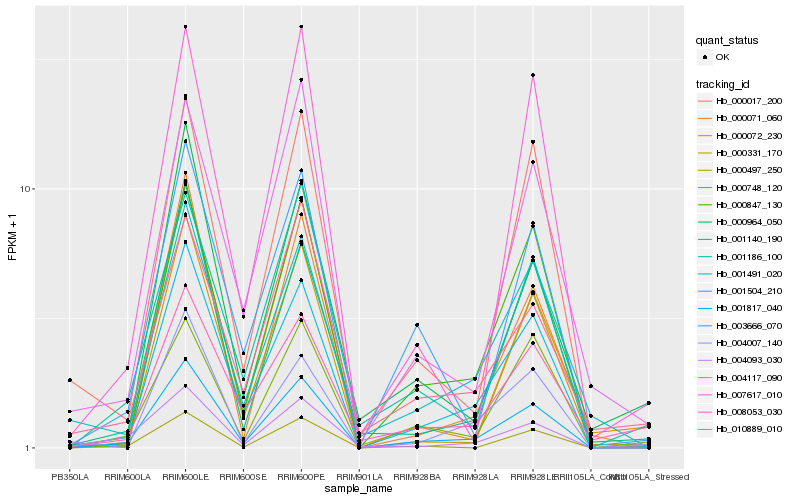
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001817_040 |
0.0 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 2 |
Hb_001504_210 |
0.0927692342 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000847_130 |
0.1205085865 |
- |
- |
PREDICTED: fimbrin-2 [Jatropha curcas] |
| 4 |
Hb_001140_190 |
0.1247596479 |
- |
- |
PREDICTED: A-kinase anchor protein 9 [Jatropha curcas] |
| 5 |
Hb_000748_120 |
0.1306746519 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL1 [Jatropha curcas] |
| 6 |
Hb_000071_060 |
0.1373763965 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004007_140 |
0.1394263289 |
- |
- |
PREDICTED: cyclin-D5-1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000497_250 |
0.140735587 |
- |
- |
PREDICTED: potassium transporter 5 [Jatropha curcas] |
| 9 |
Hb_000017_200 |
0.1410546013 |
- |
- |
PREDICTED: uncharacterized protein LOC105645766 [Jatropha curcas] |
| 10 |
Hb_003666_070 |
0.1414532925 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g66560 [Jatropha curcas] |
| 11 |
Hb_001186_100 |
0.1416143946 |
- |
- |
PREDICTED: uncharacterized protein LOC105640806 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004093_030 |
0.1454560619 |
- |
- |
hypothetical protein RCOM_1406750 [Ricinus communis] |
| 13 |
Hb_010889_010 |
0.1493696508 |
- |
- |
PREDICTED: myosin-12 [Jatropha curcas] |
| 14 |
Hb_001491_020 |
0.1495642458 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_000331_170 |
0.1497380526 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At3g03770 [Jatropha curcas] |
| 16 |
Hb_007617_010 |
0.1500749698 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 17 |
Hb_008053_030 |
0.1501617929 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 18 |
Hb_000964_050 |
0.1502739431 |
- |
- |
PREDICTED: uncharacterized protein LOC105628694 [Jatropha curcas] |
| 19 |
Hb_004117_090 |
0.1510352237 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 70 [Jatropha curcas] |
| 20 |
Hb_000072_230 |
0.1513874381 |
- |
- |
hypothetical protein POPTR_0004s10790g [Populus trichocarpa] |