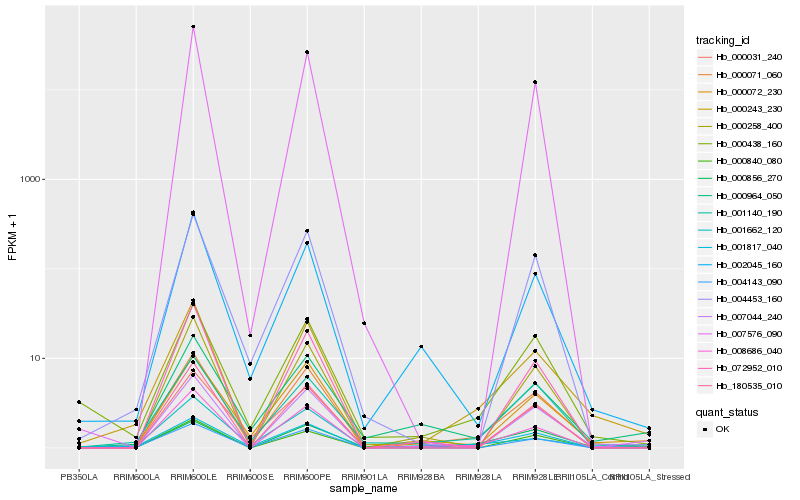
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000243_230 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_000071_060 |
0.1039120992 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_008686_040 |
0.1354778877 |
- |
- |
PREDICTED: uncharacterized protein LOC105632278 isoform X3 [Jatropha curcas] |
| 4 |
Hb_001140_190 |
0.1357569364 |
- |
- |
PREDICTED: A-kinase anchor protein 9 [Jatropha curcas] |
| 5 |
Hb_001662_120 |
0.1500739824 |
- |
- |
PREDICTED: ammonium transporter 3 member 1-like [Jatropha curcas] |
| 6 |
Hb_000072_230 |
0.1547498283 |
- |
- |
hypothetical protein POPTR_0004s10790g [Populus trichocarpa] |
| 7 |
Hb_000438_160 |
0.1547867693 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_007044_240 |
0.1588489391 |
- |
- |
PREDICTED: uncharacterized protein LOC105638704 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002045_160 |
0.1602797494 |
- |
- |
PREDICTED: DJ-1 protein homolog E-like [Jatropha curcas] |
| 10 |
Hb_000856_270 |
0.1607316837 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.6 [Jatropha curcas] |
| 11 |
Hb_000964_050 |
0.1615362505 |
- |
- |
PREDICTED: uncharacterized protein LOC105628694 [Jatropha curcas] |
| 12 |
Hb_004143_090 |
0.1649618803 |
- |
- |
PREDICTED: VIN3-like protein 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000840_080 |
0.1652871428 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 9 [Jatropha curcas] |
| 14 |
Hb_001817_040 |
0.1655602238 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 15 |
Hb_000258_400 |
0.1658965491 |
transcription factor |
TF Family: bHLH |
transcription regulator, putative [Ricinus communis] |
| 16 |
Hb_007576_090 |
0.1675644836 |
- |
- |
lipid transfer protein precursor [Gossypium herbaceum subsp. africanum] |
| 17 |
Hb_004453_160 |
0.1683369101 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_180535_010 |
0.1695479653 |
- |
- |
- |
| 19 |
Hb_000031_240 |
0.1705488007 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 20 |
Hb_072952_010 |
0.1708260517 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g18430-like [Populus euphratica] |