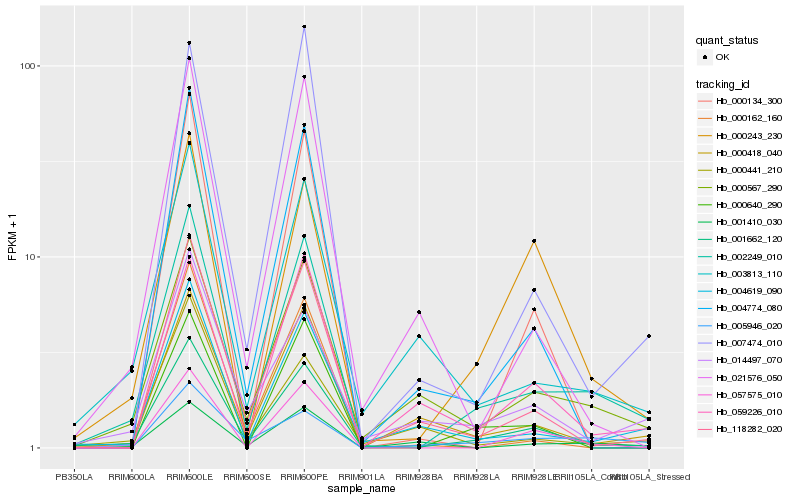
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002249_010 |
0.0 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 2 |
Hb_001662_120 |
0.1122179638 |
- |
- |
PREDICTED: ammonium transporter 3 member 1-like [Jatropha curcas] |
| 3 |
Hb_000567_290 |
0.1517583913 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 4 |
Hb_004619_090 |
0.1619013049 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003813_110 |
0.1683337364 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000418_040 |
0.1773392055 |
- |
- |
hypothetical protein RCOM_0377590 [Ricinus communis] |
| 7 |
Hb_118282_020 |
0.1775967248 |
- |
- |
PREDICTED: uncharacterized protein LOC105631532 isoform X3 [Jatropha curcas] |
| 8 |
Hb_000243_230 |
0.1858113359 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_014497_070 |
0.1869403326 |
- |
- |
PREDICTED: cyclin-dependent kinase B2-2 [Jatropha curcas] |
| 10 |
Hb_004774_080 |
0.1876293346 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_057575_010 |
0.1903781736 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000640_290 |
0.1913125227 |
- |
- |
basic helix-loop-helix protein [Populus simonii x Populus nigra] |
| 13 |
Hb_001410_030 |
0.1928053092 |
- |
- |
hypothetical protein JCGZ_20419 [Jatropha curcas] |
| 14 |
Hb_021576_050 |
0.1943681716 |
- |
- |
PREDICTED: early nodulin-like protein 1 [Jatropha curcas] |
| 15 |
Hb_007474_010 |
0.1967418121 |
- |
- |
vacuolar invertase [Manihot esculenta] |
| 16 |
Hb_005946_020 |
0.1981923905 |
- |
- |
Pleiotropic drug resistance protein 4 [Triticum urartu] |
| 17 |
Hb_059226_010 |
0.2013643792 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 18 |
Hb_000134_300 |
0.2025053925 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_000162_160 |
0.2040665256 |
- |
- |
PREDICTED: auxin transporter-like protein 5 [Populus euphratica] |
| 20 |
Hb_000441_210 |
0.2064353974 |
- |
- |
PREDICTED: uncharacterized protein LOC105636842 [Jatropha curcas] |