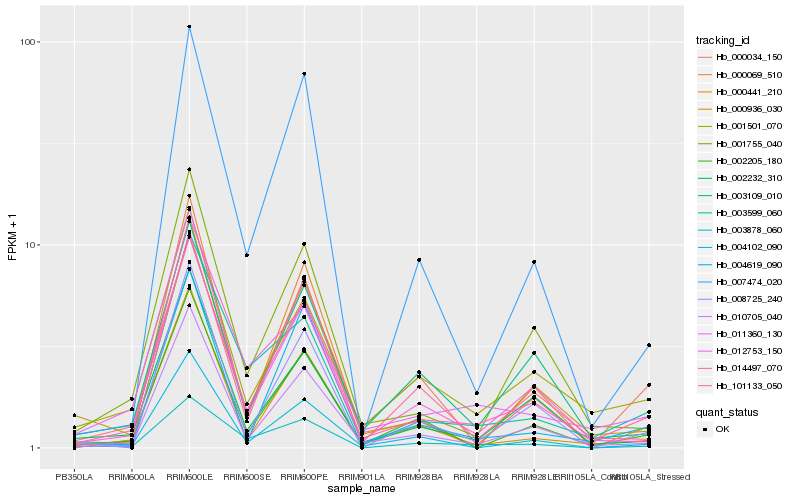
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014497_070 |
0.0 |
- |
- |
PREDICTED: cyclin-dependent kinase B2-2 [Jatropha curcas] |
| 2 |
Hb_010705_040 |
0.1034584561 |
- |
- |
PREDICTED: kinesin-4 [Jatropha curcas] |
| 3 |
Hb_011360_130 |
0.1090124613 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001755_040 |
0.1198891021 |
- |
- |
PREDICTED: cyclin-A2-4-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_002205_180 |
0.1200300324 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 6 |
Hb_002232_310 |
0.1242341378 |
- |
- |
PREDICTED: uncharacterized protein LOC105635987 [Jatropha curcas] |
| 7 |
Hb_000441_210 |
0.1256735198 |
- |
- |
PREDICTED: uncharacterized protein LOC105636842 [Jatropha curcas] |
| 8 |
Hb_001501_070 |
0.1276133077 |
- |
- |
PREDICTED: condensin complex subunit 2 [Jatropha curcas] |
| 9 |
Hb_007474_020 |
0.1319338294 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 10 |
Hb_003599_060 |
0.1323882541 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 11 |
Hb_004102_090 |
0.1364794612 |
transcription factor |
TF Family: CPP |
hypothetical protein JCGZ_06186 [Jatropha curcas] |
| 12 |
Hb_101133_050 |
0.1366319202 |
- |
- |
PREDICTED: condensin complex subunit 3 [Jatropha curcas] |
| 13 |
Hb_000936_030 |
0.1369585827 |
- |
- |
hypothetical protein RCOM_0615680 [Ricinus communis] |
| 14 |
Hb_012753_150 |
0.1385793973 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 15 |
Hb_003878_060 |
0.1397940435 |
- |
- |
hypothetical protein B456_007G326900, partial [Gossypium raimondii] |
| 16 |
Hb_003109_010 |
0.1398015164 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 17 |
Hb_004619_090 |
0.1419272358 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_008725_240 |
0.1422865741 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000034_150 |
0.1423579463 |
- |
- |
PREDICTED: kinesin-like protein BC2 [Jatropha curcas] |
| 20 |
Hb_000069_510 |
0.1434710094 |
- |
- |
PREDICTED: histone H3-like centromeric protein HTR12 [Jatropha curcas] |