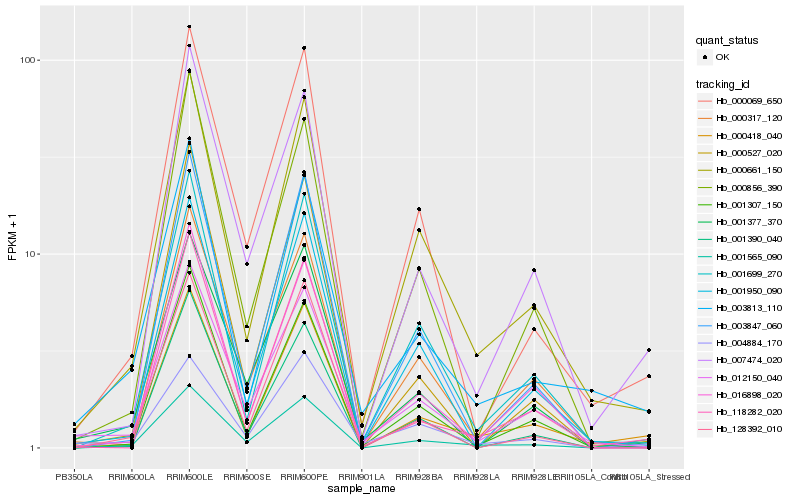
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001565_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637840 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000069_650 |
0.1023375757 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000661_150 |
0.1033821083 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 4 |
Hb_000418_040 |
0.1052611033 |
- |
- |
hypothetical protein RCOM_0377590 [Ricinus communis] |
| 5 |
Hb_007474_020 |
0.1141410607 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 6 |
Hb_001377_370 |
0.1202284263 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 7 |
Hb_001390_040 |
0.1231183689 |
- |
- |
PREDICTED: uncharacterized protein LOC105635987 [Jatropha curcas] |
| 8 |
Hb_001699_270 |
0.1242091669 |
- |
- |
PREDICTED: uncharacterized protein LOC105628996 [Jatropha curcas] |
| 9 |
Hb_001307_150 |
0.1254528394 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 10 |
Hb_003847_060 |
0.1276639793 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 11 |
Hb_012150_040 |
0.1295364766 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_118282_020 |
0.1298093905 |
- |
- |
PREDICTED: uncharacterized protein LOC105631532 isoform X3 [Jatropha curcas] |
| 13 |
Hb_128392_010 |
0.1316528487 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein At3g19184 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001950_090 |
0.1354294996 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1C [Jatropha curcas] |
| 15 |
Hb_000856_390 |
0.1367958156 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004884_170 |
0.1370218863 |
- |
- |
hypothetical protein AALP_AA1G067000 [Arabis alpina] |
| 17 |
Hb_000317_120 |
0.1371218808 |
- |
- |
PREDICTED: uncharacterized protein LOC105647000 [Jatropha curcas] |
| 18 |
Hb_000527_020 |
0.1393964789 |
- |
- |
PREDICTED: uncharacterized protein LOC105649490 [Jatropha curcas] |
| 19 |
Hb_016898_020 |
0.1401444924 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105120899 [Populus euphratica] |
| 20 |
Hb_003813_110 |
0.1408747135 |
- |
- |
conserved hypothetical protein [Ricinus communis] |