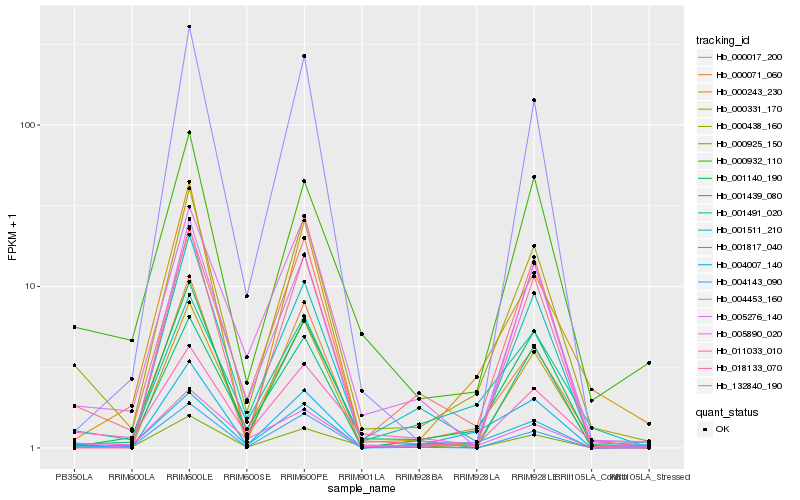
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000438_160 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001140_190 |
0.1176564169 |
- |
- |
PREDICTED: A-kinase anchor protein 9 [Jatropha curcas] |
| 3 |
Hb_000017_200 |
0.1229986568 |
- |
- |
PREDICTED: uncharacterized protein LOC105645766 [Jatropha curcas] |
| 4 |
Hb_000331_170 |
0.1353006077 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At3g03770 [Jatropha curcas] |
| 5 |
Hb_000071_060 |
0.136853224 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000932_110 |
0.1401725185 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 7 |
Hb_001491_020 |
0.1422322718 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_004007_140 |
0.1428271098 |
- |
- |
PREDICTED: cyclin-D5-1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_004143_090 |
0.1451459561 |
- |
- |
PREDICTED: VIN3-like protein 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005276_140 |
0.1452721646 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001439_080 |
0.1507676546 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 12 |
Hb_001511_210 |
0.1509346405 |
- |
- |
PREDICTED: protein ESKIMO 1-like [Jatropha curcas] |
| 13 |
Hb_132840_190 |
0.1522898794 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_011033_010 |
0.1524051713 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 15 |
Hb_005890_020 |
0.1528926936 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 16 |
Hb_004453_160 |
0.1533309146 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001817_040 |
0.1547736665 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 18 |
Hb_000243_230 |
0.1547867693 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_000925_150 |
0.1551332332 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 20 |
Hb_018133_070 |
0.155560131 |
- |
- |
PREDICTED: RING-H2 finger protein ATL43 [Jatropha curcas] |