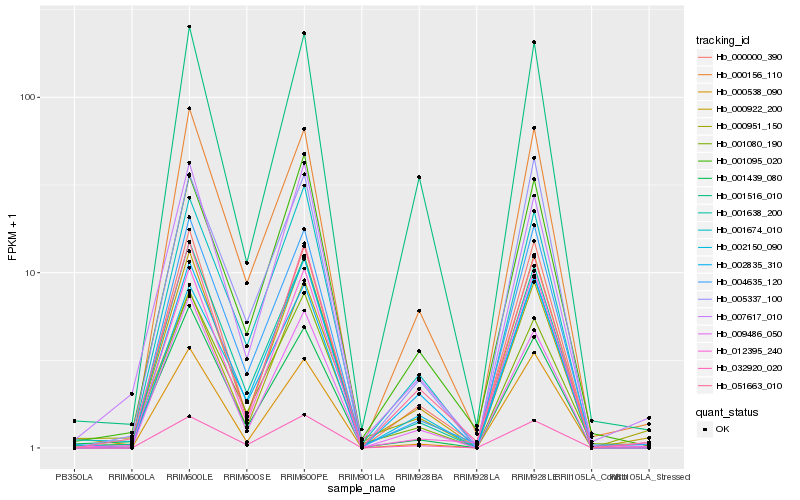
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000922_200 |
0.0 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0012s08280g [Populus trichocarpa] |
| 2 |
Hb_000951_150 |
0.0704809146 |
- |
- |
hypothetical protein POPTR_0015s05850g [Populus trichocarpa] |
| 3 |
Hb_051663_010 |
0.0790403575 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 4 |
Hb_000000_390 |
0.0793629232 |
- |
- |
PREDICTED: uncharacterized protein LOC105633753 [Jatropha curcas] |
| 5 |
Hb_004635_120 |
0.0841286756 |
- |
- |
PREDICTED: rho GTPase-activating protein 4-like [Jatropha curcas] |
| 6 |
Hb_001638_200 |
0.0879172104 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 43 [Jatropha curcas] |
| 7 |
Hb_012395_240 |
0.0884387377 |
- |
- |
PREDICTED: uncharacterized protein LOC105638421 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000538_090 |
0.0905630315 |
transcription factor |
TF Family: NAC |
hypothetical protein RCOM_1603380 [Ricinus communis] |
| 9 |
Hb_032920_020 |
0.0908644218 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 10 |
Hb_001516_010 |
0.0910260922 |
- |
- |
PREDICTED: guanylate kinase 2 [Jatropha curcas] |
| 11 |
Hb_000156_110 |
0.0954580882 |
- |
- |
Uncharacterized protein TCM_029490 [Theobroma cacao] |
| 12 |
Hb_001674_010 |
0.0978289178 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 13 |
Hb_001080_190 |
0.0996028099 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_009486_050 |
0.1008305085 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |
| 15 |
Hb_001439_080 |
0.1029558271 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 16 |
Hb_005337_100 |
0.1041422342 |
- |
- |
PREDICTED: uncharacterized protein LOC105629349 [Jatropha curcas] |
| 17 |
Hb_007617_010 |
0.1042579773 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002835_310 |
0.1049909275 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_001095_020 |
0.1061569721 |
- |
- |
PREDICTED: F-box protein At2g26850-like [Jatropha curcas] |
| 20 |
Hb_002150_090 |
0.1061750571 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |