| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
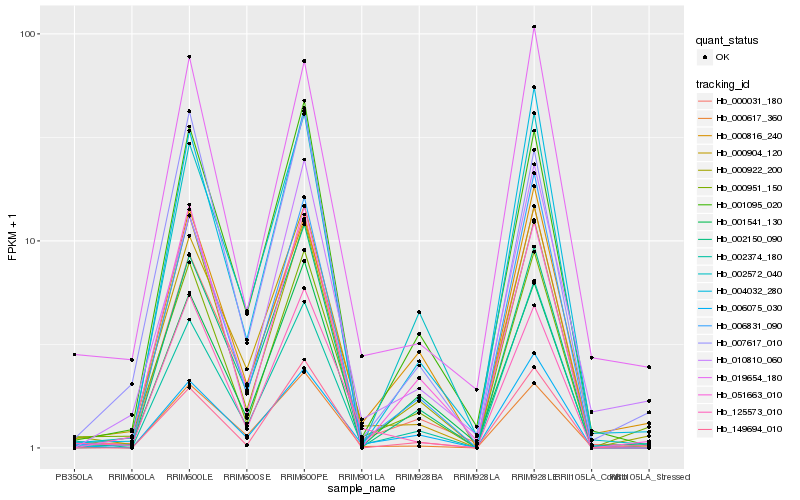
Hb_000951_150 |
0.0 |
- |
- |
hypothetical protein POPTR_0015s05850g [Populus trichocarpa] |
| 2 |
Hb_000922_200 |
0.0704809146 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0012s08280g [Populus trichocarpa] |
| 3 |
Hb_019654_180 |
0.1111474091 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 4 |
Hb_051663_010 |
0.1126047422 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 5 |
Hb_000816_240 |
0.1132141188 |
- |
- |
PREDICTED: phosphoglucan phosphatase LSF2, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000904_120 |
0.1157850777 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_149694_010 |
0.118496361 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_001541_130 |
0.1203171563 |
- |
- |
GDSL esterase/lipase [Morus notabilis] |
| 9 |
Hb_002374_180 |
0.1203336948 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 10 |
Hb_006831_090 |
0.1208087784 |
- |
- |
PREDICTED: uncharacterized protein LOC105642125 isoform X1 [Jatropha curcas] |
| 11 |
Hb_007617_010 |
0.1214455916 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002150_090 |
0.1214903421 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 13 |
Hb_125573_010 |
0.1217103999 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 14 |
Hb_004032_280 |
0.1225482909 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |
| 15 |
Hb_000617_360 |
0.1231689134 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001095_020 |
0.1238013424 |
- |
- |
PREDICTED: F-box protein At2g26850-like [Jatropha curcas] |
| 17 |
Hb_000031_180 |
0.1250074809 |
- |
- |
sarcosine oxidase, putative [Ricinus communis] |
| 18 |
Hb_002572_040 |
0.1259438514 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |
| 19 |
Hb_006075_030 |
0.12605907 |
- |
- |
PREDICTED: uncharacterized protein LOC105641890 isoform X1 [Jatropha curcas] |
| 20 |
Hb_010810_060 |
0.1262539279 |
- |
- |
sterol desaturase, putative [Ricinus communis] |