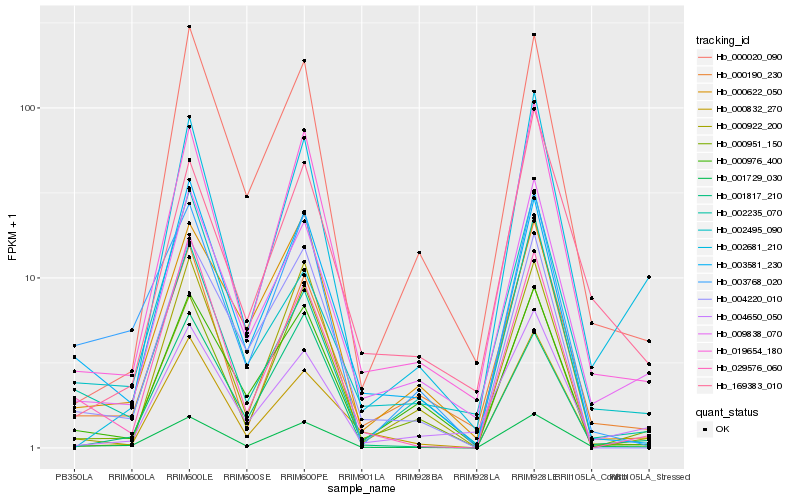
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001729_030 |
0.0 |
- |
- |
hypothetical protein VITISV_044457 [Vitis vinifera] |
| 2 |
Hb_019654_180 |
0.0956535237 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 3 |
Hb_009838_070 |
0.1185733639 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003768_020 |
0.1268030874 |
- |
- |
PREDICTED: uncharacterized protein LOC105644157 [Jatropha curcas] |
| 5 |
Hb_000190_230 |
0.1295117915 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 6 |
Hb_003581_230 |
0.1319414903 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SNF1-related protein kinase regulatory subunit gamma-1-like [Jatropha curcas] |
| 7 |
Hb_029576_060 |
0.138019617 |
- |
- |
PREDICTED: uncharacterized protein LOC105646366 [Jatropha curcas] |
| 8 |
Hb_000832_270 |
0.1390958524 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000951_150 |
0.1411831529 |
- |
- |
hypothetical protein POPTR_0015s05850g [Populus trichocarpa] |
| 10 |
Hb_004220_010 |
0.1476234902 |
- |
- |
PREDICTED: uncharacterized protein LOC105642366 [Jatropha curcas] |
| 11 |
Hb_002681_210 |
0.1496029733 |
- |
- |
PREDICTED: uncharacterized protein LOC105633686 [Jatropha curcas] |
| 12 |
Hb_001817_210 |
0.1502616603 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000020_090 |
0.1528082903 |
- |
- |
hypothetical protein JCGZ_09084 [Jatropha curcas] |
| 14 |
Hb_004650_050 |
0.1541116721 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_002235_070 |
0.156180995 |
- |
- |
PREDICTED: uncharacterized protein LOC105644108 [Jatropha curcas] |
| 16 |
Hb_000922_200 |
0.1565662268 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0012s08280g [Populus trichocarpa] |
| 17 |
Hb_000622_050 |
0.1591917626 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000976_400 |
0.1602623986 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 19 |
Hb_002495_090 |
0.1621807658 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 20 |
Hb_169383_010 |
0.162854141 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |