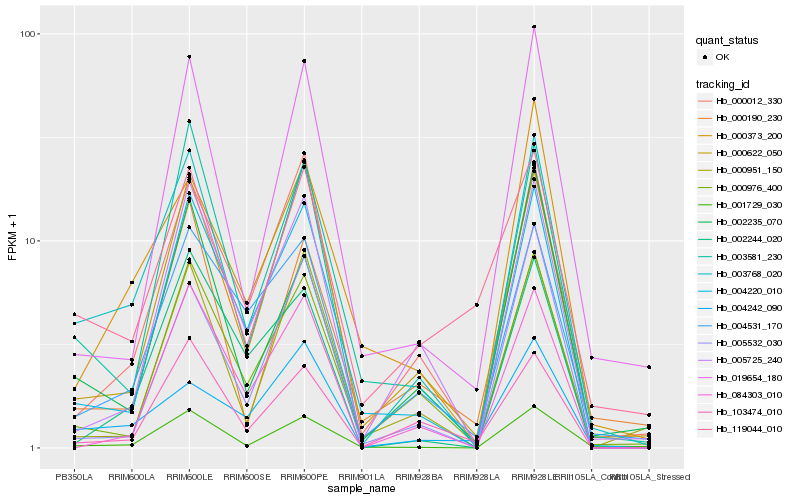
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003768_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644157 [Jatropha curcas] |
| 2 |
Hb_001729_030 |
0.1268030874 |
- |
- |
hypothetical protein VITISV_044457 [Vitis vinifera] |
| 3 |
Hb_003581_230 |
0.1452268201 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SNF1-related protein kinase regulatory subunit gamma-1-like [Jatropha curcas] |
| 4 |
Hb_004220_010 |
0.1503423936 |
- |
- |
PREDICTED: uncharacterized protein LOC105642366 [Jatropha curcas] |
| 5 |
Hb_000622_050 |
0.15428607 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000012_330 |
0.1543312149 |
- |
- |
pollen specific protein sf21, putative [Ricinus communis] |
| 7 |
Hb_000976_400 |
0.1581718538 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 8 |
Hb_002235_070 |
0.1639052557 |
- |
- |
PREDICTED: uncharacterized protein LOC105644108 [Jatropha curcas] |
| 9 |
Hb_005725_240 |
0.1645871149 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH74 isoform X1 [Jatropha curcas] |
| 10 |
Hb_019654_180 |
0.1648028165 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 11 |
Hb_004531_170 |
0.1707845826 |
- |
- |
PREDICTED: probable inactive nicotinamidase At3g16190 [Jatropha curcas] |
| 12 |
Hb_000373_200 |
0.171639696 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Populus euphratica] |
| 13 |
Hb_000190_230 |
0.174352905 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 14 |
Hb_004242_090 |
0.1760788703 |
- |
- |
hypothetical protein JCGZ_00767 [Jatropha curcas] |
| 15 |
Hb_002244_020 |
0.1802565167 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |
| 16 |
Hb_000951_150 |
0.1802830897 |
- |
- |
hypothetical protein POPTR_0015s05850g [Populus trichocarpa] |
| 17 |
Hb_084303_010 |
0.1807524215 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_103474_010 |
0.181226459 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 19 |
Hb_119044_010 |
0.1814389233 |
- |
- |
PREDICTED: stomatal closure-related actin-binding protein 2-like [Jatropha curcas] |
| 20 |
Hb_005532_030 |
0.1823187734 |
- |
- |
DNA binding protein, putative [Ricinus communis] |