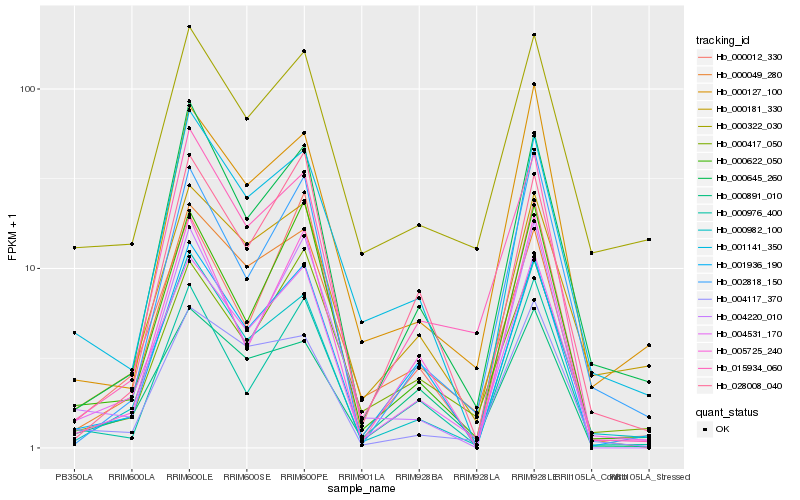
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004531_170 |
0.0 |
- |
- |
PREDICTED: probable inactive nicotinamidase At3g16190 [Jatropha curcas] |
| 2 |
Hb_000622_050 |
0.0949258215 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000982_100 |
0.0953497105 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor PIF1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001936_190 |
0.1033190442 |
- |
- |
hypothetical protein JCGZ_17780 [Jatropha curcas] |
| 5 |
Hb_004117_370 |
0.1143548484 |
- |
- |
PREDICTED: U-box domain-containing protein 16 [Jatropha curcas] |
| 6 |
Hb_000012_330 |
0.1145184879 |
- |
- |
pollen specific protein sf21, putative [Ricinus communis] |
| 7 |
Hb_000181_330 |
0.1160054845 |
- |
- |
ubiquitin ligase protein cop1, putative [Ricinus communis] |
| 8 |
Hb_000417_050 |
0.1179482561 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase MRH1 [Jatropha curcas] |
| 9 |
Hb_000645_260 |
0.1200881762 |
- |
- |
PREDICTED: uncharacterized protein LOC105643508 [Jatropha curcas] |
| 10 |
Hb_000976_400 |
0.1209987469 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 11 |
Hb_000049_280 |
0.1226538838 |
- |
- |
PREDICTED: ribokinase-like isoform X4 [Jatropha curcas] |
| 12 |
Hb_002818_150 |
0.1228011259 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0014s10680g [Populus trichocarpa] |
| 13 |
Hb_005725_240 |
0.1240874712 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH74 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000322_030 |
0.1249656211 |
- |
- |
PREDICTED: uncharacterized protein LOC105637911 [Jatropha curcas] |
| 15 |
Hb_001141_350 |
0.1267364641 |
- |
- |
hypothetical protein JCGZ_02174 [Jatropha curcas] |
| 16 |
Hb_000127_100 |
0.1271429264 |
- |
- |
PREDICTED: myb-related transcription factor, partner of profilin-like [Populus euphratica] |
| 17 |
Hb_028008_040 |
0.1279657733 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000891_010 |
0.1301041225 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004220_010 |
0.1318580944 |
- |
- |
PREDICTED: uncharacterized protein LOC105642366 [Jatropha curcas] |
| 20 |
Hb_015934_060 |
0.1335195185 |
- |
- |
PREDICTED: mitogen-activated protein kinase 20 [Jatropha curcas] |