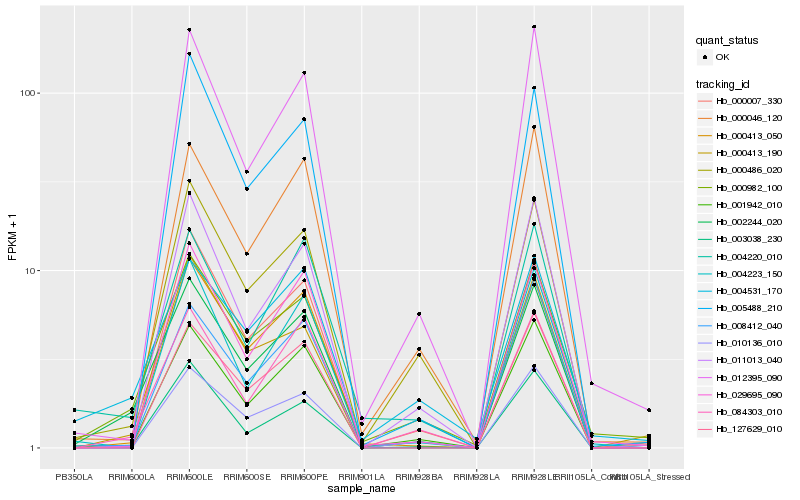
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002244_020 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |
| 2 |
Hb_000982_100 |
0.1031516057 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor PIF1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_001942_010 |
0.1049596758 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 4 |
Hb_084303_010 |
0.1135286531 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000413_050 |
0.1178872489 |
- |
- |
PREDICTED: transcription factor bHLH112 isoform X2 [Jatropha curcas] |
| 6 |
Hb_003038_230 |
0.1234518998 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 7 [Jatropha curcas] |
| 7 |
Hb_000486_020 |
0.1270693314 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.2 [Jatropha curcas] |
| 8 |
Hb_004220_010 |
0.1282016858 |
- |
- |
PREDICTED: uncharacterized protein LOC105642366 [Jatropha curcas] |
| 9 |
Hb_011013_040 |
0.1324993134 |
transcription factor |
TF Family: C2C2-YABBY |
unknown [Populus trichocarpa] |
| 10 |
Hb_029695_090 |
0.1332076072 |
- |
- |
PREDICTED: probable inactive patatin-like protein 9 [Jatropha curcas] |
| 11 |
Hb_012395_090 |
0.1337845229 |
- |
- |
PREDICTED: protein PROTON GRADIENT REGULATION 5, chloroplastic [Jatropha curcas] |
| 12 |
Hb_010136_010 |
0.1346732375 |
- |
- |
Putative receptor-like protein kinase [Glycine soja] |
| 13 |
Hb_127629_010 |
0.1348480133 |
- |
- |
hypothetical protein CICLE_v10005216mg [Citrus clementina] |
| 14 |
Hb_008412_040 |
0.1356698852 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_000413_190 |
0.1363013182 |
- |
- |
PREDICTED: oxygen-evolving enhancer protein 3-2, chloroplastic [Pyrus x bretschneideri] |
| 16 |
Hb_000007_330 |
0.1381742055 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 17 |
Hb_004223_150 |
0.1385459522 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Jatropha curcas] |
| 18 |
Hb_004531_170 |
0.1386556069 |
- |
- |
PREDICTED: probable inactive nicotinamidase At3g16190 [Jatropha curcas] |
| 19 |
Hb_000046_120 |
0.1389709244 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 20 |
Hb_005488_210 |
0.1417470059 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |