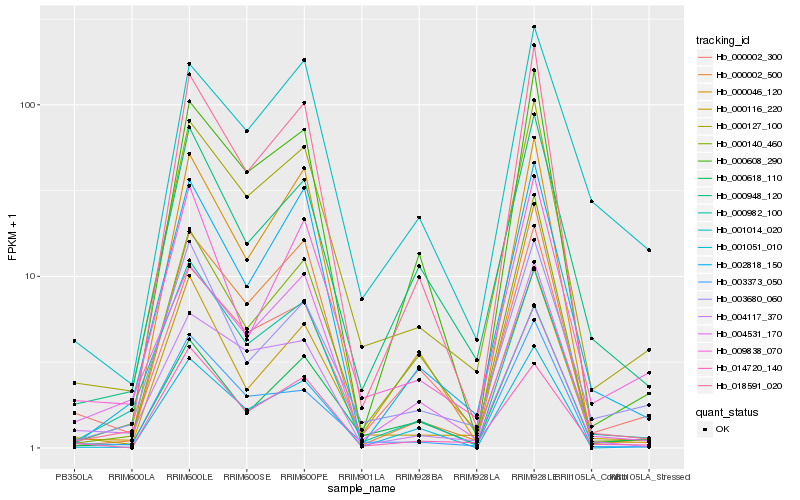
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000127_100 |
0.0 |
- |
- |
PREDICTED: myb-related transcription factor, partner of profilin-like [Populus euphratica] |
| 2 |
Hb_000002_500 |
0.1108856196 |
- |
- |
PREDICTED: uncharacterized protein LOC105632094 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000116_220 |
0.1157606411 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000982_100 |
0.1160906079 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor PIF1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000618_110 |
0.1168384788 |
- |
- |
hypothetical protein POPTR_0003s09870g [Populus trichocarpa] |
| 6 |
Hb_002818_150 |
0.1176644168 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0014s10680g [Populus trichocarpa] |
| 7 |
Hb_004117_370 |
0.1195764695 |
- |
- |
PREDICTED: U-box domain-containing protein 16 [Jatropha curcas] |
| 8 |
Hb_000608_290 |
0.1211317468 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 9 |
Hb_003680_060 |
0.1227601085 |
- |
- |
hypothetical protein VITISV_022322 [Vitis vinifera] |
| 10 |
Hb_000002_300 |
0.1228105623 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 11 |
Hb_000140_460 |
0.1230089764 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 12 |
Hb_001051_010 |
0.1236730792 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |
| 13 |
Hb_000948_120 |
0.1242119413 |
- |
- |
PREDICTED: uncharacterized protein LOC105634688 isoform X1 [Jatropha curcas] |
| 14 |
Hb_018591_020 |
0.1252113023 |
- |
- |
Ribulose bisphosphate carboxylase/oxygenase activase 1, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_014720_140 |
0.126295063 |
- |
- |
PREDICTED: uncharacterized protein LOC105640367 [Jatropha curcas] |
| 16 |
Hb_009838_070 |
0.1266608379 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001014_020 |
0.1270298367 |
- |
- |
gene GA family protein [Populus trichocarpa] |
| 18 |
Hb_004531_170 |
0.1271429264 |
- |
- |
PREDICTED: probable inactive nicotinamidase At3g16190 [Jatropha curcas] |
| 19 |
Hb_000046_120 |
0.1284123321 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 20 |
Hb_003373_050 |
0.1295246603 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |