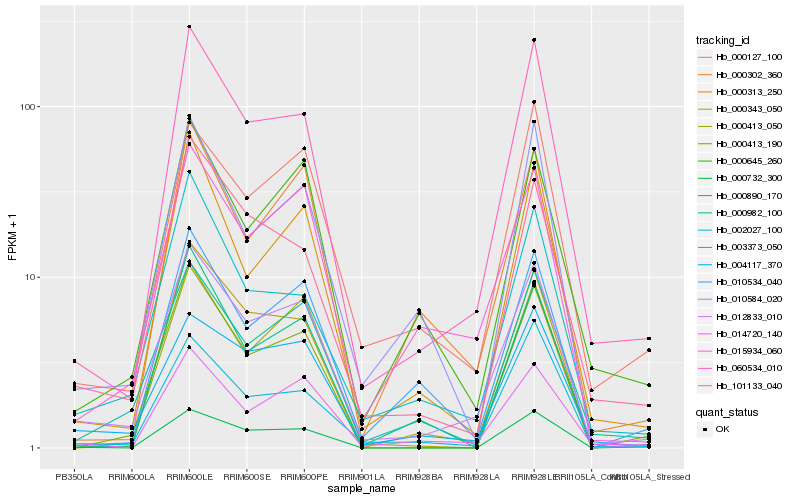
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012833_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 2 |
Hb_060534_010 |
0.0967411716 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 3 |
Hb_004117_370 |
0.1079323991 |
- |
- |
PREDICTED: U-box domain-containing protein 16 [Jatropha curcas] |
| 4 |
Hb_014720_140 |
0.1109399868 |
- |
- |
PREDICTED: uncharacterized protein LOC105640367 [Jatropha curcas] |
| 5 |
Hb_015934_060 |
0.1111060606 |
- |
- |
PREDICTED: mitogen-activated protein kinase 20 [Jatropha curcas] |
| 6 |
Hb_000982_100 |
0.1192601821 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor PIF1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000313_250 |
0.1334133241 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 8 |
Hb_000343_050 |
0.134735142 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000645_260 |
0.1350021055 |
- |
- |
PREDICTED: uncharacterized protein LOC105643508 [Jatropha curcas] |
| 10 |
Hb_101133_040 |
0.1365767256 |
- |
- |
PREDICTED: nudix hydrolase 18, mitochondrial-like [Jatropha curcas] |
| 11 |
Hb_000127_100 |
0.1381966392 |
- |
- |
PREDICTED: myb-related transcription factor, partner of profilin-like [Populus euphratica] |
| 12 |
Hb_002027_100 |
0.140322732 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000413_190 |
0.1403474514 |
- |
- |
PREDICTED: oxygen-evolving enhancer protein 3-2, chloroplastic [Pyrus x bretschneideri] |
| 14 |
Hb_000732_300 |
0.1412664 |
- |
- |
PREDICTED: uncharacterized protein LOC105649465 isoform X2 [Jatropha curcas] |
| 15 |
Hb_010534_040 |
0.1414098812 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000302_360 |
0.1446352294 |
- |
- |
beta-carotene hydroxylase, putative [Ricinus communis] |
| 17 |
Hb_010584_020 |
0.1454158273 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 18 |
Hb_000413_050 |
0.1457730356 |
- |
- |
PREDICTED: transcription factor bHLH112 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003373_050 |
0.14732075 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 20 |
Hb_000890_170 |
0.1473958186 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |