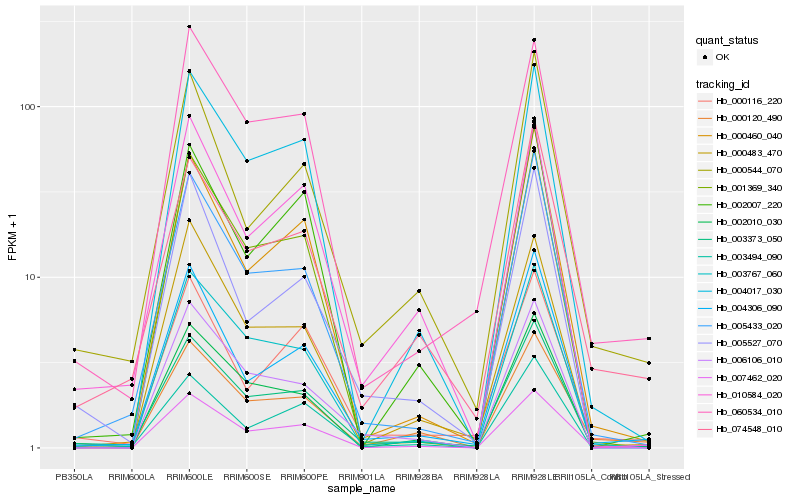
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003373_050 |
0.0 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 2 |
Hb_005433_020 |
0.0714033098 |
- |
- |
PREDICTED: multicopper oxidase LPR1 [Jatropha curcas] |
| 3 |
Hb_004306_090 |
0.0928454149 |
- |
- |
PREDICTED: uncharacterized protein LOC105644392 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002010_030 |
0.1020548616 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 5 |
Hb_003767_060 |
0.1043833091 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 6 |
Hb_000460_040 |
0.1045534785 |
- |
- |
PREDICTED: protein phosphatase 2C 57 [Jatropha curcas] |
| 7 |
Hb_006106_010 |
0.1065891196 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_007462_020 |
0.1076932818 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 9 |
Hb_060534_010 |
0.1079920393 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 10 |
Hb_005527_070 |
0.1082229018 |
- |
- |
short-chain dehydrogenase/reductase family protein [Populus trichocarpa] |
| 11 |
Hb_004017_030 |
0.1082407011 |
- |
- |
PREDICTED: 2-hydroxyisoflavanone dehydratase-like [Jatropha curcas] |
| 12 |
Hb_000544_070 |
0.1088911228 |
- |
- |
Peptidyl-prolyl cis-trans isomerase FKBP17-2 [Morus notabilis] |
| 13 |
Hb_074548_010 |
0.1090455085 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_010584_020 |
0.1112315715 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 15 |
Hb_003494_090 |
0.1140751367 |
transcription factor |
TF Family: MYB |
myb family transcription factor family protein [Populus trichocarpa] |
| 16 |
Hb_000120_490 |
0.1157669523 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3-like [Jatropha curcas] |
| 17 |
Hb_000116_220 |
0.1161328374 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000483_470 |
0.1165506499 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_001369_340 |
0.1173431292 |
- |
- |
Peroxisomal membrane protein 11B [Theobroma cacao] |
| 20 |
Hb_002007_220 |
0.1178965652 |
- |
- |
PREDICTED: signal recognition particle 43 kDa protein, chloroplastic [Jatropha curcas] |