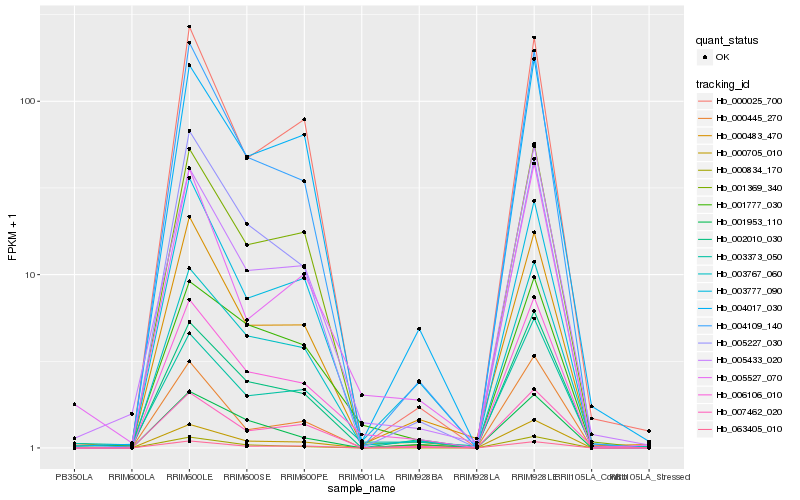
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006106_010 |
0.0 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_003767_060 |
0.081521056 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 3 |
Hb_002010_030 |
0.0942157383 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 4 |
Hb_004109_140 |
0.0960154828 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001369_340 |
0.1011819704 |
- |
- |
Peroxisomal membrane protein 11B [Theobroma cacao] |
| 6 |
Hb_000483_470 |
0.1027345783 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_005433_020 |
0.1060108826 |
- |
- |
PREDICTED: multicopper oxidase LPR1 [Jatropha curcas] |
| 8 |
Hb_001777_030 |
0.1061662902 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 9 |
Hb_003373_050 |
0.1065891196 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 10 |
Hb_007462_020 |
0.1087510161 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 11 |
Hb_004017_030 |
0.10967214 |
- |
- |
PREDICTED: 2-hydroxyisoflavanone dehydratase-like [Jatropha curcas] |
| 12 |
Hb_003777_090 |
0.1125537282 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 13 |
Hb_005227_030 |
0.1184086568 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP28, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001953_110 |
0.1202466972 |
- |
- |
PREDICTED: protein TAPETUM DETERMINANT 1-like [Jatropha curcas] |
| 15 |
Hb_000834_170 |
0.1204365397 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 16 |
Hb_000025_700 |
0.1220946109 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 17 |
Hb_063405_010 |
0.1233822729 |
- |
- |
PREDICTED: probable terpene synthase 3 [Jatropha curcas] |
| 18 |
Hb_000445_270 |
0.1239841299 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_000705_010 |
0.1247822369 |
- |
- |
hypothetical protein CICLE_v10027296mg [Citrus clementina] |
| 20 |
Hb_005527_070 |
0.125301757 |
- |
- |
short-chain dehydrogenase/reductase family protein [Populus trichocarpa] |