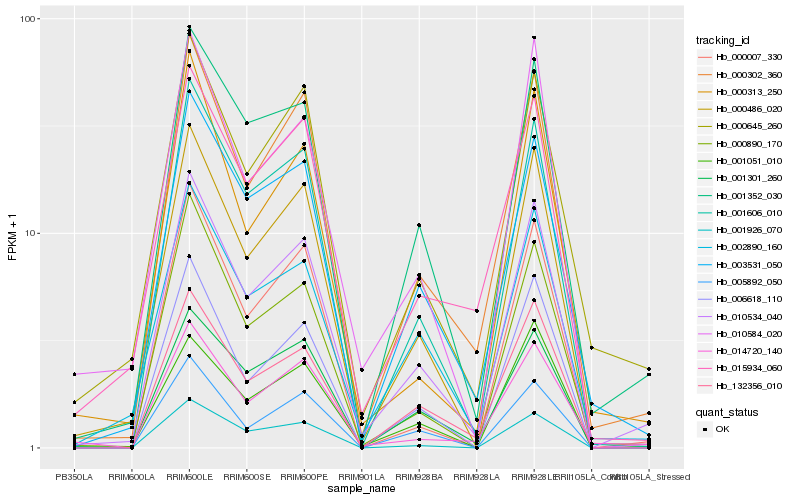
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010534_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000302_360 |
0.0707752319 |
- |
- |
beta-carotene hydroxylase, putative [Ricinus communis] |
| 3 |
Hb_000486_020 |
0.0772671732 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.2 [Jatropha curcas] |
| 4 |
Hb_001352_030 |
0.0822030516 |
- |
- |
PREDICTED: uncharacterized protein LOC105636310 [Jatropha curcas] |
| 5 |
Hb_001606_010 |
0.0877211209 |
- |
- |
kinase, putative [Ricinus communis] |
| 6 |
Hb_000645_260 |
0.0930374376 |
- |
- |
PREDICTED: uncharacterized protein LOC105643508 [Jatropha curcas] |
| 7 |
Hb_002890_160 |
0.0937055129 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000890_170 |
0.0950058718 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |
| 9 |
Hb_003531_050 |
0.0958428496 |
- |
- |
PREDICTED: pyridoxal reductase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_006618_110 |
0.0992271376 |
- |
- |
- |
| 11 |
Hb_132356_010 |
0.1000826664 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 12 |
Hb_001926_070 |
0.1049211192 |
- |
- |
hypothetical protein JCGZ_21510 [Jatropha curcas] |
| 13 |
Hb_010584_020 |
0.1053105956 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 14 |
Hb_001051_010 |
0.1095237481 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |
| 15 |
Hb_000007_330 |
0.1096143769 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 16 |
Hb_005892_050 |
0.1103524954 |
- |
- |
PREDICTED: uncharacterized protein LOC105632175 [Jatropha curcas] |
| 17 |
Hb_014720_140 |
0.1108208856 |
- |
- |
PREDICTED: uncharacterized protein LOC105640367 [Jatropha curcas] |
| 18 |
Hb_015934_060 |
0.1108981863 |
- |
- |
PREDICTED: mitogen-activated protein kinase 20 [Jatropha curcas] |
| 19 |
Hb_000313_250 |
0.1125707549 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 20 |
Hb_001301_260 |
0.1129342789 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |