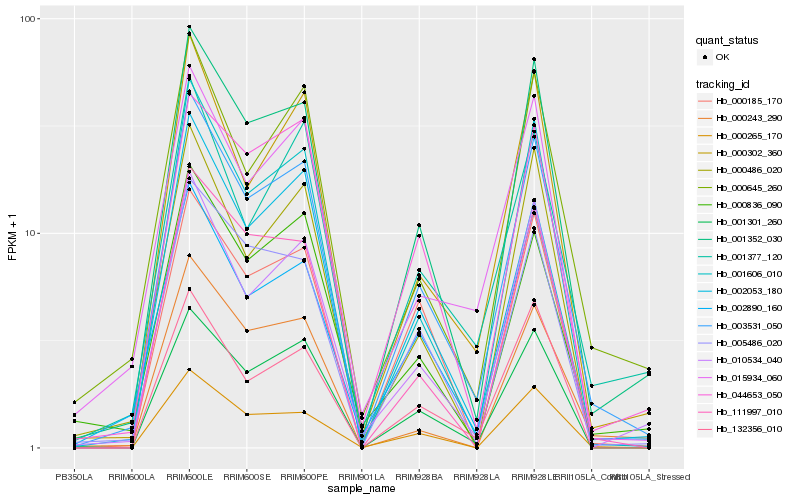
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003531_050 |
0.0 |
- |
- |
PREDICTED: pyridoxal reductase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001352_030 |
0.06173567 |
- |
- |
PREDICTED: uncharacterized protein LOC105636310 [Jatropha curcas] |
| 3 |
Hb_001606_010 |
0.0853844166 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_001301_260 |
0.0893071154 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_002890_160 |
0.0893123397 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000265_170 |
0.0924623278 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 7 |
Hb_000302_360 |
0.0926267263 |
- |
- |
beta-carotene hydroxylase, putative [Ricinus communis] |
| 8 |
Hb_010534_040 |
0.0958428496 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002053_180 |
0.0983244548 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 10 |
Hb_000645_260 |
0.0999086269 |
- |
- |
PREDICTED: uncharacterized protein LOC105643508 [Jatropha curcas] |
| 11 |
Hb_005486_020 |
0.101268085 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 12 |
Hb_015934_060 |
0.106525047 |
- |
- |
PREDICTED: mitogen-activated protein kinase 20 [Jatropha curcas] |
| 13 |
Hb_111997_010 |
0.1096167273 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Vitis vinifera] |
| 14 |
Hb_132356_010 |
0.1098535972 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 15 |
Hb_001377_120 |
0.1107670452 |
- |
- |
PREDICTED: uncharacterized protein LOC105642861 [Jatropha curcas] |
| 16 |
Hb_000836_090 |
0.1120285472 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 17 |
Hb_000185_170 |
0.1124826964 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_000486_020 |
0.1132313341 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.2 [Jatropha curcas] |
| 19 |
Hb_044653_050 |
0.1137731513 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000243_290 |
0.1148138894 |
- |
- |
plastidic aldolase family protein [Populus trichocarpa] |