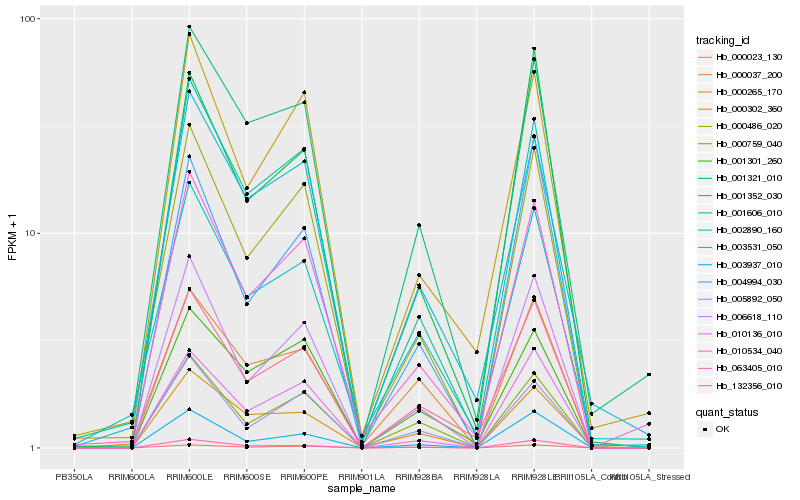
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_132356_010 |
0.0 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 2 |
Hb_000023_130 |
0.083560527 |
- |
- |
Metal-nicotianamine transporter YSL1 [Glycine soja] |
| 3 |
Hb_006618_110 |
0.0887298842 |
- |
- |
- |
| 4 |
Hb_000759_040 |
0.0961355333 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 5 |
Hb_001301_260 |
0.1000151759 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_010534_040 |
0.1000826664 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000302_360 |
0.1002054056 |
- |
- |
beta-carotene hydroxylase, putative [Ricinus communis] |
| 8 |
Hb_063405_010 |
0.1015290449 |
- |
- |
PREDICTED: probable terpene synthase 3 [Jatropha curcas] |
| 9 |
Hb_001606_010 |
0.1021725685 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_000486_020 |
0.105606405 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.2 [Jatropha curcas] |
| 11 |
Hb_005892_050 |
0.1084557317 |
- |
- |
PREDICTED: uncharacterized protein LOC105632175 [Jatropha curcas] |
| 12 |
Hb_001321_010 |
0.1088980213 |
- |
- |
HIGH-CHLOROPHYLL-FLUORESCENCE 101 family protein [Populus trichocarpa] |
| 13 |
Hb_002890_160 |
0.1091141181 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003937_010 |
0.109806108 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 15 |
Hb_003531_050 |
0.1098535972 |
- |
- |
PREDICTED: pyridoxal reductase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001352_030 |
0.1106515068 |
- |
- |
PREDICTED: uncharacterized protein LOC105636310 [Jatropha curcas] |
| 17 |
Hb_000265_170 |
0.112189883 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 18 |
Hb_010136_010 |
0.1134267406 |
- |
- |
Putative receptor-like protein kinase [Glycine soja] |
| 19 |
Hb_000037_200 |
0.1136991704 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_004994_030 |
0.1162172901 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |