| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
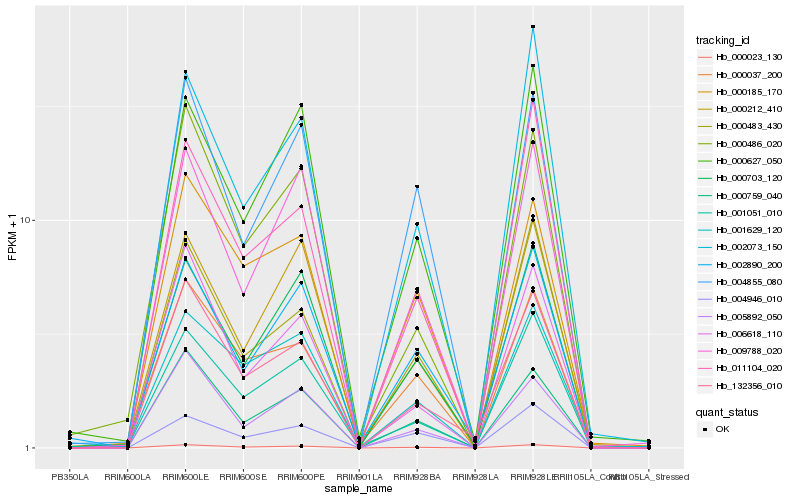
Hb_000023_130 |
0.0 |
- |
- |
Metal-nicotianamine transporter YSL1 [Glycine soja] |
| 2 |
Hb_000759_040 |
0.0706605466 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 3 |
Hb_001051_010 |
0.0830887178 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |
| 4 |
Hb_132356_010 |
0.083560527 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 5 |
Hb_009788_020 |
0.0836154771 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g06840 [Jatropha curcas] |
| 6 |
Hb_000037_200 |
0.0848528325 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_001629_120 |
0.0857701483 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Populus euphratica] |
| 8 |
Hb_000483_430 |
0.0869987164 |
- |
- |
- |
| 9 |
Hb_000703_120 |
0.0875428572 |
- |
- |
PREDICTED: uncharacterized protein LOC105635706 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000185_170 |
0.0908870833 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_006618_110 |
0.093576549 |
- |
- |
- |
| 12 |
Hb_004855_080 |
0.0946838725 |
- |
- |
hypothetical protein JCGZ_25039 [Jatropha curcas] |
| 13 |
Hb_000627_050 |
0.0956675947 |
- |
- |
PREDICTED: uncharacterized protein LOC105632845 [Jatropha curcas] |
| 14 |
Hb_004946_010 |
0.0978121736 |
- |
- |
hypothetical protein POPTR_0013s035502g, partial [Populus trichocarpa] |
| 15 |
Hb_002073_150 |
0.0978936716 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 16 |
Hb_011104_020 |
0.0991225142 |
- |
- |
PREDICTED: ABC transporter B family member 2-like [Jatropha curcas] |
| 17 |
Hb_005892_050 |
0.099804416 |
- |
- |
PREDICTED: uncharacterized protein LOC105632175 [Jatropha curcas] |
| 18 |
Hb_000486_020 |
0.1009678868 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.2 [Jatropha curcas] |
| 19 |
Hb_000212_410 |
0.1019161956 |
- |
- |
PREDICTED: serine carboxypeptidase 1-like [Jatropha curcas] |
| 20 |
Hb_002890_200 |
0.1024787339 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |