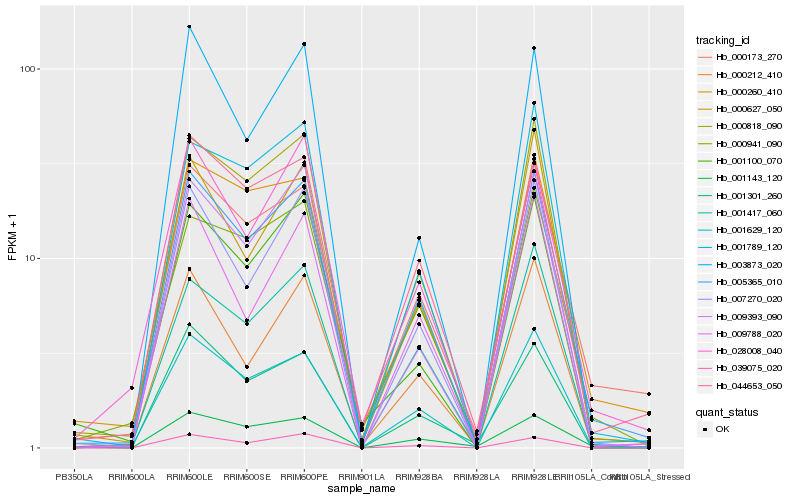
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005365_010 |
0.0 |
- |
- |
PREDICTED: serine carboxypeptidase-like 20 [Jatropha curcas] |
| 2 |
Hb_001629_120 |
0.0508319832 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Populus euphratica] |
| 3 |
Hb_000173_270 |
0.0762123847 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000818_090 |
0.0782637574 |
- |
- |
glutamate receptor 3 plant, putative [Ricinus communis] |
| 5 |
Hb_007270_020 |
0.0808656329 |
- |
- |
hypothetical protein JCGZ_09593 [Jatropha curcas] |
| 6 |
Hb_001417_060 |
0.0911246744 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 7 |
Hb_044653_050 |
0.0914306289 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 8 |
Hb_009393_090 |
0.0919235592 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000212_410 |
0.094258603 |
- |
- |
PREDICTED: serine carboxypeptidase 1-like [Jatropha curcas] |
| 10 |
Hb_039075_020 |
0.095914577 |
- |
- |
PREDICTED: uncharacterized protein LOC103331245 [Prunus mume] |
| 11 |
Hb_001789_120 |
0.0962441399 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_000941_090 |
0.097252096 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 13 |
Hb_028008_040 |
0.0973152119 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000260_410 |
0.0983375202 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |
| 15 |
Hb_000627_050 |
0.0992631447 |
- |
- |
PREDICTED: uncharacterized protein LOC105632845 [Jatropha curcas] |
| 16 |
Hb_001301_260 |
0.1001433222 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_001100_070 |
0.1041861162 |
- |
- |
PREDICTED: ferredoxin--nitrite reductase, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_003873_020 |
0.1063529316 |
- |
- |
PREDICTED: root phototropism protein 2 [Jatropha curcas] |
| 19 |
Hb_009788_020 |
0.1066684942 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g06840 [Jatropha curcas] |
| 20 |
Hb_001143_120 |
0.1073966278 |
- |
- |
PREDICTED: uncharacterized protein LOC105638955 [Jatropha curcas] |